
- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons

Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

Determine Structure with Combined Spectra
- Last updated
- Save as PDF
- Page ID 167069

- Chris Schaller
- College of Saint Benedict/Saint John's University
There are a many ways we can use NMR spectroscopy to analyse compounds. One common application is in determination of an unknown structure. Given the MS, IR, 13 C and 1 H NMR spectra, what might be the structure of an unknown sample?
It is often easiest to start with the IR spectrum.
- identify at least three peaks in the IR spectrum. Which peaks seem to tell you the most information about this compound?
- don't think with your head; think with your hands. Write down ideas on the spectrum.
- if you are working on a formal proof of structure, on a class test or a lab report, you may be required to enter your data in a table correlating wavenumber with peak assignment:
For example, a student might obtain the following IR spectrum.
From that information, she constructs the following table. She might even write this table, by hand, directly on her spectrum. She makes useful notes on the edges, and might even include some guesses, which she later crosses out, but does not erase. She is assisted in this task by consulting an IR table, that suggests what some of these peaks might mean.

- make special note of what atoms are present in the compound: C, H, N, O...
- also note your initial ideas about specific functional groups that may be present.
- if you are unsure of an assignment, put a question mark beside it to signal this uncertainty.
- some data may need to be discarded later if it is not consistent with other data.
Look at the 13 C spectrum.
- How many different carbons are there, based on the number of peaks in the spectrum? This is the first step in estimating the molecular formula.
- Do you have reason to believe there is symmetry in the structure? In the entire compound or just part of it? Adjust the number of carbons you think you are dealing with.
- As in IR spectroscopy, begin assigning peaks, either on the spectrum or, if required, in a table:
For example, a student might obtain the following 13 C NMR spectrum:
From that information, she puts together the following table:

- you will be able to assign all peaks in the NMR spectrum, not just a few like in IR.
You can get more information on the formula from the 1 H NMR spectrum and the mass spectrum.
- In the 1 H NMR spectrum, what does the sum of the integrations suggest about the number of hydrogens?
- At this point you may have a formula, C x H y O z . What would be the mass of a compound with this formula?
- Compare this mass to the mass spectrum. Does it match?
- If not, consider whether a common atom (such as oxygen) is missing from your formula, or if there might be symmetry that you have missed.
- Does the mass spectrum suggest the presence of a nitrogen (odd molecular weight) or chlorine or bromine (isotope pattern)?
Once you have the molecular formula, the number of possible structures is automatically limited. The number of units of unsaturation can help you narrow down the possibilities.
- The ratio of C : H in a saturated, acyclic hydrocarbon is n : 2n+2.
- Each pair of H missing from the formula corresponds to a multiple bond or a ring.
- The presence of oxygen (which is divalent) does not alter the C : H ratio.
- The presence of halogen (which is monovalent) means there is one H missing.
- The presence of a nitrogen (which is trivalent) means there is an extra H in the formula.
- In other words, C 4 H 8 O has one ring or double bond just like C 4 H 8 , and so do C 4 H 7 Br and C 4 H 9 N.
As in 13 C NMR, you should be able to assign all peaks in the 1 H NMR spectrum. You may be able to do so by making notes on the spectrum. If you think you know the structure, you may be able to draw it and note which peak belongs with which proton.
A formal proof of structure might require a table of assignments.
- This table demonstrates your ability to read the spectrum. Can you decide what ratio of protons is suggested by the integral line? Can you decide whether a peak is a quartet?
- The partial structure column should explain the shift, integration and multiplicity for the peak in that row. It should not show any other information from elsewhere in the structure. This restriction forces you to demonstrate a thorough understanding of the data in a way that "getting the right answer" does not.
- The partial structure column is best filled in with drawings, not words. The drawing is a partial structure.
- Because the partial structure will show the protons absorbing at the shift in that row as well the neighbouring protons, you need to distinguish between them in your picture. Most people circle or underline or make bold the protons that show up at the shift given in that row.
- When finished with the partial structure column, you should be able to link the partial structures together to make an entire structure in the assignment column.
An example of a spectrum and its accompanying data table is given below. Here is the spectrum:
Here is a data table:
Things to note:
- This student has used two integration columns instead of just one.
- The first column shows the integral measured from the spectrum. She probably used a ruler.
- The second column, which she called int(n), contains a convenient ratio taken from the raw data. This ratio is easier to use in her assignments.
- Also note that the peak at 9.7 ppm does not have a very good integral. There is either a "phasing" or a "level & tilt" problem here that can be corrected using the NMR software, but this is sometimes difficult to do. If she had taken an automatic printout of this integral measurement, she would have gotten a strange number; in this case, it would be about -5, because the end of the integral line is lower than the start. It clearly isn't a negative number of hydrogens, though. She has instead measured the vertical rise in the integral and recorded that; it isn't perfect, but is a fair estimate in this case.
Contributors and Attributions
Chris P Schaller, Ph.D. , (College of Saint Benedict / Saint John's University)
Pep-Calc.com: a set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment
- Open access
- Published: 24 February 2016
- Volume 30 , pages 271–277, ( 2016 )
Cite this article
You have full access to this open access article

- Sam Lear 1 &
- Steven L. Cobb 1
7259 Accesses
63 Citations
2 Altmetric
Explore all metrics
The ability to calculate molecular properties such as molecular weights, isoelectric points, and extinction coefficients is vital for scientists using and/or synthesizing peptides and peptoids for research. A suite of two web utilities: Peptide Calculator and Peptoid Calculator, available free at http://www.pep-calc.com , are presented. Both tools allow the calculation of peptide/peptoid chemical formulae and molecular weight, ChemDraw structure file export and automatic assignment of mass spectral peaks to deletion sequences and metal/protecting group adducts. Peptide Calculator also provides a calculated isoelectric point, molar extinction coefficient, graphical peptide charge summary and β -strand contiguity profile (for aggregation-prone sequences), indicating potential regions of synthesis difficulty. In addition to the unique automatic spectral assignment features offered across both utilities, Peptoid Calculator represents a first-of-a-kind resource for researchers in the field of peptoid science. With a constantly expanding database of over 120 amino acids, non-natural peptide building blocks and peptoid building blocks, it is anticipated that Pep-Calc.com will act as a valuable asset to those working on the synthesis and/or application of peptides and peptoids in the biophysical and life sciences fields.
Similar content being viewed by others
pyPept: a python library to generate atomistic 2D and 3D representations of peptides
MassSpecBlocks: a web-based tool to create building blocks and sequences of nonribosomal peptides and polyketides for tandem mass spectra analysis
PEPCONF, a diverse data set of peptide conformational energies
Avoid common mistakes on your manuscript.
Introduction
Convenient and rapid access to calculated molecular properties is essential for researchers using and/or synthesizing peptides and peptidomimetics for biophysical or life sciences applications. Furthermore, the process of assigning peptide byproducts in mass spectra resulting from residue deletions or incomplete protecting group removal during a synthesis can be a laborious and time consuming process, and access to freely available automatic assignment tools is necessary to improve workflow and increase research efficiency. While a plethora of peptide and protein property calculation tools are accessible online, very few offer mass spectral peak assignment functionality, and for those that do this is often extremely limited.
While the ExPASy portal [ 1 ] acts as the most comprehensive protein property calculation resource for molecular biology, other more specific tools exist, such as ChemCalc [ 2 ], PredictProtein [ 3 ], IMSPeptider [ 4 ], POTAMOS [ 5 ], Top Pred [ 6 ], CheckMyMetal [ 7 ], AFAL [ 8 ] and a host of other peptide property calculation utilities [ 9 – 16 ]. Few of these are designed specifically with the synthetic peptide chemist in mind however, and furthermore, to the best of our knowledge, no freely available web services exist for the calculation of peptoid molecular properties or assignment of peptoid synthesis mass spectra.
We present a pair of web tools: Peptide Calculator and Peptoid Calculator, for chemical formula and molecular weight calculation of peptides and peptoids. In addition, both sites offer automatic assignment of mass spectral peaks to deletion sequences, metal ion adducts and protected byproducts, as well as the option to download structures in ChemDraw format for the sequences entered. Peptide Calculator can also give calculated values for isoelectric point and molar extinction coefficient (at 280 nm), as well as a plot of calculated β -strand propensity for the sequence. Both utilities are available at http://www.pep-calc.com .
Features summary
Sequence input.
Peptide and peptoid sequences up to 150 residues in length can be entered, containing any combination of amino acids or peptoid building blocks present in the database. For peptides, the input string may include any of the standard single-letter amino acid codes in addition to a number of ‘nonstandard’ residues (such as phosphoserine, pS ), which must appear in parentheses within the string. An equivalent set of single-letter codes does not exist for peptoid building blocks, therefore Peptoid Calculator instead accepts a string of residue codes separated by dashes, without the requirement for multiple-letter codes to be enclosed in brackets. As peptoids can often consist of repeating motifs, Peptoid Calculator additionally allows parentheses to be used to indicate repeat sequences within the input string. Peptide and peptoid sequence input options are summarized in Fig. 1 .
Summary of input options available for Peptide Calculator and Peptoid Calculator. Sequences can be specified using a large variety of residue types, and Peptoid Calculator also accepts input strings containing repeating sequence motifs indicated by nested parentheses . Termini formulae can be selected from available options and are also fully customizable. Optionally, m / z values can be specified for automatic peak assignment
Both utilities also offer the option of specifying formulae for the N- and C-termini of the input sequence. These can be entered as a custom molecular formula string, or selected from lists of predefined formulae (Fig. 1 ). A full list of available residue types (showing residue code, molecular formula and molecular structure) and predefined termini available on Peptide Calculator and Peptoid Calculator is given on each site’s Help page.
A final (optional) input field can be used to specify m / z values belonging to singly-charged species in mass spectra, for automatic assignment to peptide or peptoid deletion sequences and/or adducts (described below).
Calculated parameters
Both utilities will provide a molecular formula and calculated molecular weight for peptide/peptoid sequences entered, in addition to an automatically generated ChemDraw structure in .cdxml format (Fig. 2 ). A spectral assignment for the peptide/peptoid will also be given if m / z values were provided as part of the input.
Both Peptide Calculator and Peptoid Calculator will output a number of basic calculated properties, in addition to a peak assignment and ChemDraw structure file for the sequence. A number of additional parameters are also provided for peptides, including estimated isoelectric point and molar extinction coefficient, as well as a graphical residue charge summary and β -strand contiguity profile
An example of an automatic peak assignment is illustrated in Fig. 3 (assignment output shown in Table 1 ). A number of peaks are present in the spectrum and have been assigned to either deletion sequences (where one or more residues are missing from the target sequence), sequences with unremoved protecting groups, metal adducts or a combination of two or more of the conditions described. Peptide Calculator and Peptoid Calculator will attempt to assign any m / z values provided to either the target sequence or a formula containing single or multiple residue deletions, metals, unremoved protecting groups or any combination thereof.
Example spectrum automatically assigned by Peptide Calculator (assignment is also available for Peptoid Calculator). A number of single- and multiple-residue deletions have been identified, in combination with sodiation and/or unremoved 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf) protecting groups. The ethyl 3-mercaptopropionate thioester is available as a predefined C-terminus and can be selected during sequence input
A number of calculated parameters specific to peptides are also available. Peptide Calculator will provide estimated values for sequence isoelectric point and molar extinction coefficient (at 280 nm), as well as a pie chart summarizing proportions of acidic, basic and uncharged residues in the sequence (Fig. 2 ). For sequences that are 10 residues or longer in length, a β -strand contiguity profile is calculated (Fig. 2 ). This provides an ab initio prediction of the location of β -strand forming regions within the sequence, and hence may offer an indication of aggregation-prone sequences, or those which are likely to present difficulties during synthesis.
Peptide Calculator and Peptoid Calculator make use of a database each containing either amino acids or peptoid building blocks defined by residue codes (single- or multiple-letter) and accompanying molecular formulae. Molecular weights are calculated by reference to a table of atomic masses (most abundant isotope). Methods used to generate other calculated parameters are described below. All Pep-Calc.com functionality is scripted using an extensible framework written in the Python programming language, and the site is accessed using an HTML web interface. Residue formulae can be added to either database upon request.
Isoelectric point and molar extinction coefficient calculation
Theoretical peptide isoelectric points are calculated using the bisection method described by Kozlowski [ 17 – 19 ]. The net charge of the peptide can be found using the Henderson–Hasselbalch equation, taking into account contributions from negatively and positively charged groups (first and second terms in Eq. ( 1 ) respectively, where K a is the acid dissociation constant of the amino acid).
As the isoelectric point (pI) represents the pH at which the net charge of the peptide equals zero, finding the root of this equation (in this case numerically, using the bisection method) gives the pI (or pH at zero charge).
Peptide Calculator takes into account side chain charge contributions from Arg, Asp, Cys, Glu, His, Lys and Tyr residues, in addition to the N-terminal amine and C-terminal carboxyl groups (only if the terminus types are set to ‘Unmodified’ and ‘Acid’ respectively). Other residue side chains are not taken into account for pI estimation, and are designated ‘Other’ in the charge summary pie chart.
Molar extinction coefficients are estimated using Eq. ( 2 ), described by Pace et al. [ 20 ]. The formula takes into account numbers of Trp and Tyr residues in the peptide ( \(n_{Trp}\) and \(n_{Tyr}\) respectively), in addition to the number of cystine residues ( \(n_{cystine}\) ) formed via disulfide bond formation between pairs of cysteine side chains (reduced cysteine residues do not contribute significantly to the absorbance above 275 nm [ 20 ]).
Peptide Calculator outputs two values for \(\varepsilon _{280}\) , calculating the theoretical molar extinction coefficient based on either formation of the maximum number of disulfide bonds possible ( \(n_{cystine}\) equal to the number of cysteine residue pairs ), or complete reduction resulting in the absence of disulfides ( \(n_{cystine} = 0\) ).
Automatic mass spectral peak assignment
User-entered m / z values are assigned through the process summarized in the flowchart given in Fig. 4 . Pep-Calc first compiles lists of possible single-amino-acid deletions and single modifications (metal adducts and unremoved protecting groups), including null entries for no deletion or no modification. A complete set of combinations of these lists is then generated, and the molecular weight of the peptide/peptoid sequence incorporating each combination of single deletion and/or single modification calculated. Each input peak is then compared against the list of molecular weights, and a peak is assigned to a particular peptide if it falls within ±1.0 u of the calculated molecular weight of the peptide.
In the event that all the input peaks are not assigned on the first pass, Pep-Calc calculates the molecular weights for all peptide/deletion/modification combinations incorporating single or double deletions and single or double modifications, and checks remaining peaks against these (omitting already assigned peaks). This process is repeated until all peaks are assigned, or until up to the maximum allowed number of deletions/modifications have been checked. To prevent excessive computation times the maximum number of deletions/modification depends on the sequence length, and is set at 5 iterations for sequences up to 30 residues in length, 4 for 60-mers and 3 up to the maximum 150 residue sequence input.
Flowchart summarizing the mass spectral peak assignment algorithm used by Peptide Calculator and Peptoid Calculator. Residues missing from the expected full sequence are termed ‘deletions’ and any other atom or group that causes a change in the molecular weight of the sequence (including metal adducts and unremoved protecting groups) is termed a ‘modification’. Which deletions and modifications are allowed depends on the residues present in the sequence (unremoved Pbf protecting groups, for example, are only permitted for Arg residues). Only sequences bearing a single deletion and/or a single modification are considered on the first iteration (N = 1), increasing to two of each on the second etc. The maximum allowed value for N depends on the length of the input sequence, and is set at 5 iterations for sequences up to and including 30 residues in length, 4 up to 60 residues and 3 up to the maximum 150 residues
Calculation of sequence β -strand propensity
β -Strand contiguity profiles for peptides greater than 9 residues in length are calculated using an implementation of the simple algorithm for sliding averages (SALSA) described by Zibaee et al. [ 21 ]. A window of size 4 residues is scanned across the input sequence and each fragment within the window scored using Eq. ( 3 ), where \(P_{\alpha },\, P_{\beta }\) and \(P_{t}\) are the Chou–Fasman secondary structure probability parameters (for α -helix, β -strand and reverse turn preference, respectively) [ 22 ]. This process is repeated for all window sizes up to 20 residues or the sequence length (whichever is reached first), and all fragments with scores lower than 1.2 are discarded.
β -Strand propensity values are then calculated for each residue in the sequence by summing the scores of all remaining windows which contain the residue. These final values are then plotted to produce a β -strand contiguity profile for the peptide. Chou–Fasman parameters are only available for the 20 canonical amino acids and hence only these are taken into account when calculating β -strand propensity values.
It should be noted that β -strand propensity alone may not be indicative of aggregation likelihood or sequence difficulty. In addition, ab initio secondary structure prediction methods based on probability parameters alone can in some cases give false predictions or fail to predict regions of a given secondary structure. SALSA was chosen with speed in mind, and for this reason the calculated profile is intended to serve only as a guide.
Conclusions
Peptide Calculator and Peptoid Calculator form a set of full featured, freely available web utilities for peptide and peptoid molecular property calculation and mass spectral peak assignment. Modern peptide research demands tools that can handle residue types beyond the canonical amino acids (such as phosphorylated peptide building blocks [ 23 – 25 ]), and with unique spectral assignment capabilities and an expanding amino acid database Peptide Calculator offers a service beyond that of current freely available web utilities. Furthermore, similar services for peptoid research are non-existent, and Peptoid Calculator represents a first-of-a-kind resource for researchers in the field of peptoid science. The tools described have found broad application in our lab, and are used frequently in peptide and peptoid research activities [ 26 – 28 ]. It is anticipated that Pep-Calc.com ( http://www.pep-calc.com ) will act as a valuable asset to those synthesizing and/or using peptides or peptoids as part of their research in the biophysical and life sciences fields.
ExPASy: SIB Bioinformatics Resource Portal. http://www.expasy.org . Accessed 03 Dec 2015
Patiny L, Borel A (2013) ChemCalc: a building block for tomorrow’s chemical infrastructure. J Chem Inf Model 53:1223–1228
Article CAS Google Scholar
Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Hönigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B (2014) PredictProtein—an open resource for online prediction of protein structural and functional features. Nucl Acids Res 42:W337–W343
de Carvalho RV, Lopez-Ferrer D, Guimarães KS, Lins RD (2013) IMSPeptider: a computational peptide collision cross-section area calculator based on a novel molecular dynamics simulation protocol. J Comput Chem 34:1707–1718
Article Google Scholar
Vlachopanos A, Soupsana E, Politou AS, Papamokos GV (2014) POTAMOS mass spectrometry calculator: computer aided mass spectrometry to the post-translational modifications of proteins. A focus on histones. Comput Biol Med 55:36–41
Claros MG, von Heijne G (1994) TopPred II: An improved software for membrane protein structure predictions. Comput Appl Biosci 10:685–686
CAS Google Scholar
Zheng H, Chordia MD, Cooper DR, Chruszcz M, Müller P, Sheldrick GM, Minor W (2014) Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc 9:156–170
Arenas-Salinas M, Ortega-Salazar S, Gonzales-Nilo F, Pohl E, Holmes DS, Quatrini R (2014) AFAL: a web service for profiling amino acids surrounding ligands in proteins. J Comput Aided Mol Des 28:1069–1076
Peptide Mass Calculator. http://immweb.vet.uu.nl/P&P_fac/pepcalc.htm . Accessed 04 Dec 2015
Peptide Property Calculator. http://www.basic.northwestern.edu/biotools/proteincalc.html . Accessed 04 Dec 2015
Fragment Ion Calculator. http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html . Accessed 04 Dec 2015
N2.cz Peptide Calculator. http://pept.n2.cz/ . Accessed 04 Dec 2015
Oligonucleotide- and Peptide calculations. http://www.chemie.hu-berlin.de/seitz/oligo-tools_e.htm . Accessed 04 Dec 2015
Sheffield ChemPuter. http://winter.group.shef.ac.uk/chemputer/ . Accessed 04 Dec 2015
PepCalc.com—Innovagen peptide property calculator. http://pepcalc.com/ . Accessed 22 Jan 2016
GenScript Peptide Property Calculator. https://www.genscript.com/ssl-bin/site2/peptide_calculation.cgi/ . Accessed 22 Jan 2016
Kozlowski LP (2015) Calculation of protein isoelectric point. http://isoelectric.ovh.org . Accessed 23 Sept 2015
Cameselle JC, Ribeiro JM, Sillero A (1986) Derivation and use of a formula to calculate the net charge of acid-base compounds. Its application to amino acids, proteins and nucleotides. Biochem Educ 14:131–136
Sillero A, Maldonado A (2006) Isoelectric point determination of proteins and other macromolecules: oscillating method. Comput Biol Med 36:157–166
Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4:2411–2423
Zibaee S, Makin OS, Goedert M, Serpell LC (2007) A simple algorithm locates β -strands in the amyloid fibril core of α -synuclein, A β , and tau using the amino acid sequence alone. Protein Sci 16:906–918
Chou PY, Fasman GD (1974) Conformational parameters for amino acids in helical, β -sheet, and random coil regions calculated from proteins. Biochemistry 13:211–222
Chan C-F, Lan R, Tsang M-K, Zhou D, Lear S, Chan W-L, Cobb SL, Wong W-K, Hao J, Wong W-T, Wong K-L (2015) Directional Plk1 inhibition-driven cell cycle interruption using amphiphilic thin-coated peptide-lanthanide upconversion nanomaterials as in vivo tumor suppressors. J Mater Chem B 3:2624–2634
Chan C-F, Xie C, Tsang M-K, Lear S, Dai L, Zhou Y, Cicho J, Karbowiak M, Hreniak D, Lan R, Cobb SL, Lam MH-W, Hao J, Wong K-L (2015) The effects of morphology and linker length on the properties of peptide-lanthanide upconversion nanomaterials as G2 phase cell cycle inhibitors. Eur J Inorg Chem 2015:4539–4545. doi: 10.1002/ejic.201500321
Li H, Chan C-F, Chan W-L, Lear S, Cobb SL, Mak N-K, Lau TC, Lan R, Wong W-K, Wong K-L (2014) Monitoring and inhibition of Plk1: amphiphilic porphyrin conjugated Plk1 specific peptides for its imaging and anti-tumor function. Org Biomol Chem 12:5876–5882
Bolt HL, Cobb SL (2016) A practical method for the synthesis of peptoids containing both lysine-type and arginine-type monomers. Org Biomol Chem 14:1211–1215
Eggimann GA, Bolt HL, Denny PW, Cobb SL (2015) Investigating the anti-leishmanial effects of linear peptoids. ChemMedChem 10:233–237
Eggimann GA, Sweeney K, Bolt HL, Rozatian N, Cobb SL, Denny PW (2015) The role of phosphoglycans in the susceptibility of Leishmania mexicana to the temporin family of anti-microbial peptides. Molecules 20:2775–2785
Download references
Acknowledgments
The authors wish to thank Hannah Bolt for the compilation of peptoid building block data, and Ehmke Pohl for assistance during preparation of the manuscript.
Financial support was provided by the Engineering and Physical Sciences Research Council (EPSRC).
Author information
Authors and affiliations.
Department of Chemistry, Durham University, South Road, Durham, DH1 3LE, UK
Sam Lear & Steven L. Cobb
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Sam Lear .

Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Reprints and permissions
About this article
Lear, S., Cobb, S.L. Pep-Calc.com: a set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J Comput Aided Mol Des 30 , 271–277 (2016). https://doi.org/10.1007/s10822-016-9902-7
Download citation
Received : 11 December 2015
Accepted : 12 February 2016
Published : 24 February 2016
Issue Date : March 2016
DOI : https://doi.org/10.1007/s10822-016-9902-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Calculated properties
- Automatic mass assignment
- ChemDraw export
- Find a journal
- Publish with us
- Track your research

Special ChemDoodle Offer
Thank you for trying our ChemDoodle Web Components demos!
You have exceeded the maximum number of free transactions allowed. For unlimited access, please consider our special offer for a ChemDoodle license which includes both ChemDoodle 2D and ChemDoodle 3D . Join the hundreds of thousands of professionals and students that use ChemDoodle every day to finish their work faster and more accurately.
Demos > Simulate NMR and MS
This demo will simulate 1 H and 13 C NMR spectra, as well as the mass spectrum parent peak (isotopic distribution), of the molecule you draw in the sketcher. Click the Simulate Spectra button to simulate the spectra when you finish drawing your molecule. The spectra are interactive, so you can change their perspectives. For more simulation options, atom assignments and publishing features, please see ChemDoodle .
Get your work done with our popular desktop software.
ChemDoodle 2D
ChemDoodle 3D
This website uses cookies to improve website functionality and performance, to analyze website traffic, and to provide you with a more personalized experience. To find out more about the cookies we use, see our Privacy Policy .
- My Playlists
- Help (UoN only)
- Media Upload
- Research News
- Student News
- Classics and Archaeology
- Engineering
- Veterinary Medicine and Science
- Plants and the Soil Environment
- Social Sciences
- Student Life
- The Nottingham Experience
- Scholarships and Funding
- Arts and Education
- Science and Engineering
- Arts and Social Sciences
- Accommodation
- Careers and Employability
- Nottingham City
- Our campuses
- Support and Wellbeing
Tutorial guide to ChemDraw
Related media.
Library Subject Guides
- Subject Guides
- Structure Drawing Tools and Nomenclature
Chemistry: Structure Drawing Tools and Nomenclature
- Books and ebooks
- Dictionaries, Encyclopedias, Handbooks
- Journal Articles and Databases
- Journal Title Abbreviations
- Data and Properties
- Exam Papers (via AKO | LEARN)
- Past Tests (via AKO | LEARN)
- Products and Prices
- Safety Data Sheets
- Information Competencies for Chemistry Undergraduates (Wikibook)
- Stages in the Research Process
- Citation Styles and EndNote
- Writing Guides
- Web Lectures
- Stay Current
- For Academics
- Library Navigator
Structure Drawing Tools
- ACD/ ChemSketch Freeware Includes tools for 2D structure cleaning, 3D optimization and viewing, InChI generation and conversion, drawing of polymers, organometallics, Markush structures, and IUPAC systematic naming capability for molecules with fewer than 50 atoms and 3 rings. Free download; already installed on all computers in Chemistry Level 5 Computer Suite.
- BIOVIA Draw ( formerly Accelrys Draw and before that Symyx Draw ) Free for academic and personal use. Draw atoms and bonds, change bond order, change atom properties, create rings. Built-in structure–to–name and name–to–structure converters for IUPAC, SMILES and InChI.
- ChemSpider A free database of chemistry structures and their associated information. To draw and search on a structure or substructure, click “Structure search,” then click the pencil inside the diagram, then select “Draw or Edit”
- PubChem PubChem Structure Search allows the PubChem Compound Database to be to be queried by chemical structure or chemical structure pattern. The PubChem Sketcher allows a query to be drawn manually. Users may also specify the structural query input by PubChem Compound Identifier (CID) , SMILES , SMARTS , InChI , Molecular Formula , or by upload of a supported structure file format
- PyMOL A molecular visualisation system, maintained and distributed by Schrödinger. Free download for educational use.
- ChemDraw ChemDraw includes Struct=Name, ChemDraw/Excel and ChemNMR. Create stereochemically correct structures from chemical names, and get accurate IUPAC names for structures. Estimate NMR spectra from a ChemDraw structure with direct atom to spectral correlation. The ChemDraw ActiveX/Plugin adds chemical intelligence to your browser for querying databases and displaying information. UC has a site licence for CambridgeSoft ChemDraw Prime and PerkinElmer ChemDraw Professional for use on UC computers. Log a Ticket to ITS for it to be pushed to the Software Centre on a particular computer.
Nomenclature
- InChI Trust The InChI Trust develops and promotes the use of the IUPAC InChI open-source chemical structure representation algorithm. Its website includes videos explaining InChI and downloadable software
- IUPAC – Nomenclature (with link to IUPAC color books)
- MarvinSketch – Structure to IUPAC Name MarvinSketch is a chemical editor for drawing chemical structures, queries and reactions. Although it is the default structure editor for Reaxys, it can be downloaded for personal use. The "Traditional Name" or the "Preferred IUPAC Name" name can be found after drawing a structure, then click 'Insert' and 'IUPAC Name'
- NCI/CADD Chemical Identifier Resolver Converts structures into identifiers or identifiers (InChI and Smiles) into structures or other identifiers (e.g. IUPAC names and CAS RNs)
- OPSIN: Open Parser for Systematic IUPAC nomenclature Input a chemical name and OPSIN returns its depiction, SMILES string, InChI and its CML
- PubChem Sketcher From drawn structures generates SMILES, SMARTS, InChI and InChIKey
- << Previous: Safety Data Sheets
- Next: Syntheses >>
- Last Updated: May 8, 2024 2:29 PM
- URL: https://canterbury.libguides.com/chem
Assignments
Note: Do not forget to have a look at our 1H NMR Automatic Assignments Tutorial
Mnova provides a very simple interface to assign your molecule. Open your NMR spectrum and load a molecule structure. Then follow the menu 'Analysis/Assignment' (or use the shortcut 'A').
Click on an atom on the molecular structure (or a spectrum region) and then release the mouse and drag it to your desired peak. Once your desired peak is highlighted on the spectrum, click on it to assign it.
This peak will now be assigned to the atom (which will turn to green). Once the assignment has been made, you will get an atom number label on the chemical shift and hovering the mouse over the atom will highlight the applicable peak in the spectrum and hovering the mouse over the peak will highlight the corresponding atom on the molecular structure.
You can also assign a region of the spectrum just by clicking, dragging and releasing the mouse over the desired region. In the example below, we have assigned a -CH2 group, so a new window will be displayed to allow us to select which atom we want to assign, 18, 18', both (in blank) or even we can select any other annotation: 18a, 18b, cis/trans, ax/eq, etc:
Assign a multiplet by dragging the mouse to the 'multiplet box' (in this case the name of the multiplet is replaced with the atom number) or to the 'integral curve', as you can see in the picture below:
We recommend you to assign your atoms to your multiplet boxes in order to transfer assignments through datasets. you can also assign a 13C-NMR of the same molecule in the same document. To do that, select the atoms from the Table of molecules by following the menu 'View/Tables/Molecules'. In order to keep the assignments propagation, follow the menu 'View/Tables/Assignments' to select what datasets you want to take into account (in the example below, we have selected the 1H and the 13C NMR datasets).
Assignments to 2D spectra:
Once you have assigned the 1H and the 13C spectra, if you open in the same document a 2D-NMR spectrum and you link the spectra (from the Assignments table), you will see the assignments graphically on the screen and hovering the mouse over the atom will highlight the applicable chemical shift. In the example below, you can see the correlation between the two hydrogens and the carbon of the position 12 in a HSQC:
Also you can carry out assignments to 2D spectra by typing the applicable number of the atom in the 'Assignments Table' or graphically by selecting the atom in the molecule structure and the corresponding signal in the 2D:
Click on the 2D spectra and a new window will be displayed to select the atoms are involved. (in that case, you will only need to select the atom 10 in f1):
Finally click OK and hover the mouse over the atom 23 to see the result (please notice how the assignment table has been automatically filled with the number '10' in the HSQC column, in order to show the correlation between the C-10 and the H-23).
Copying several datasets on the same page is also available. Here you can see a page which contains a 1H, 13C and HSQC datasets with an assignment in the molecular structure:
About Author

Related Posts

Mnova System Requirements and Mbook (In-house installations)

Arithmetics

A robust, general automatic phase correction algorithm for high-resolution NMR data
Comments are closed.

- 产品 / PRODUCT
- 购买 / BUY NOW
- 下载 / DOWNLOAD
- 支持 / SUPPORT

ChemDraw 15的Structure菜单里面有什么?
发布时间:2016-03-09 10: 23: 40
ChemDraw是一款在全球使用的纯英文版软件,这也为中国用户掌握ChemDraw使用方法提高了难度。ChemDraw软件接口比较多,功能较为全面,再加上英文版本,所以掌握使用方法就更加困难。为了帮助每位ChemDraw使用者更加透彻地理解Structure菜单这个 ChemDraw 工具的功能,下文小编将具体介绍每个命令的中文含义。
Structure菜单中有什么命令?
Structure菜单是ChemDraw工具栏中的重要成员之一,熟练掌握的话大大有助于高效绘制化学结构,菜单命令有以下几个方向:
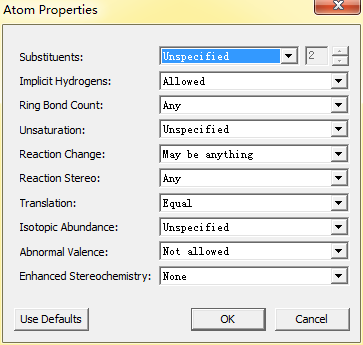
1、属性管理:AtomProperties、Bond Properties和Bracket Properties可以分别打开原子、化学键以及括号属性设置框,用户可以更改原子或化学键的属性,满足自己的绘制要求,原子属性设置框如下图所示:
2、结构式整理:Check Structure 可以快速检验结构以查出结构式是否存在问题并给出提示信息;Clean Up Structure、Clean Up Reaction和Clean Up Biopolymer分别可以整理结构式、反应式以及聚合物结构使得结构看起来更加美观;Expand Label和Contract Label分别是扩展和收缩标记;Expand Generic Structure扩展同类结构式;
3、添加结构:Add Multi-Center Attachment可以增加多中心附件;Add Variable Attachment是增加灵活可变附件;R-Logic Query是查询同类结构;Add 3D Property是增加结构的3D特性;Enhanced Stereochemistry是增强立体化学效果;
4、反应式分析:Map Reaction Atoms表示绘制对应的反应原子;Clear Reaction Map可以清除对应的反应原子;Analyze Stoichiometry是分析化学计量;
5、预测图谱:Predict 1H NMR Shift和Predict 13C NMR Shifts分别是预测1H NMR和13C NMR谱图;Make Spectrum-Structure Assignment表示分配光谱结构;
6、结构式转换:Define Nickname可以定义俗名,将常用结构定义为俗名以后便于快速绘制结构式;Convert Name to Structure是将名字转化为结构式;Convert Structureto Name是将结构式转化为名称。
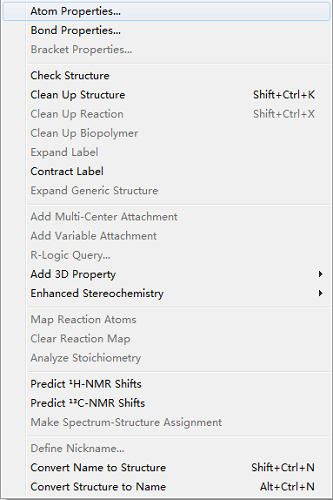
以上就是对ChemDraw 15的Structure菜单功能的介绍,功能比较多,只有熟悉每个命令才能在绘制结构时异常轻松。如果需要学习更多的ChemDraw使用技巧请点击 教你如何添加ChemDraw快捷键 。
标签: ChemDraw 15 , ChemDraw菜单 , ChemDraw使用方法
- 上一篇: ChemDraw 15菜单功能介绍(一)
- 下一篇: ChemDraw 15菜单功能介绍(二)
- # chemdraw程序无响应什么原因 chemdraw程序无响应怎么办
- # chem3d优化结构在哪里 chem3d怎么优化结构
- # chemdraw下载教程 chemdraw安装包地址
- # chemdraw画图怎么缩小 chemdraw画图工具不见了
- # chemdraw标注手性中心 chemdraw标注手性中心绝对构型
新闻期刊 | 认证激活 | 如何使用 知识储备 | 生物医药 | 材料科学 有机化学 | 新手入门 | 常见问题 应用范围 | 产品介绍 | 分析化学 高分子材料 | 化学工程 |
- chemdraw括号怎么打 chemdraw括号怎么旋转
- 分子结构绘制软件有哪些 分子结构绘制软件哪个好
- chemdraw粗黑线什么意思 chemdraw粗黑线怎么做
- chemdraw默认画布大小 chemdraw默认键长怎么调
- chemdraw默认保存地址 chemdraw默认字体设置
- ChemDraw设置默认格式方法 ChemDraw格式是什么意思
- ChemDraw工具栏使用技巧 ChemDraw工具栏怎么调节
- ChemDraw工具栏调到左边的方法 ChemDraw工具栏变为竖直的原因
- ChemDraw工具栏消失了怎么办 ChemDraw工具栏没有圆形什么原因
- ChemDraw如何设置acs格式 ChemDraw设置acs 1996方法
- chemdraw下载教程 chemdraw安装包地址
- ChemDraw怎么加甲基 ChemDraw画r基
- ChemDraw设置化学式格式 ChemDraw怎么设置默认格式
- ChemDraw如何更改键的粗细 ChemDraw设置化学式格式
- ChemDraw调整箭头大小 ChemDraw怎么让箭头边缘变粗
- ChemDraw化合物编号 ChemDraw化合物结构的粗细
- chemdraw怎么输入希腊字母及特殊字符
- chemdraw怎么输入基团 chemdraw基团在哪里
- ChemDraw画图规范 ChemDraw画图时连接处怎么画
- 怎么调整化学分子量到4位小数?
下 载 | 产 品 | 应 用 | 支 持 | 关于我们 | 网站地图 | 欢迎各位经销商洽谈合作事宜
本网站并非Revvity创建,由Revvity在中国的合作伙伴Suzhou C&J Marketing Co.,Ltd. 创建运营。可验证邮箱: [email protected]

Copyright © 2024 ChemDraw - 苏州苏杰思网络有限公司旗下网站 | 经营许可证编号:苏B1.B2-20150264 | 证照信息 | 特聘法律顾问:江苏政纬律师事务所 宋红波

IMAGES
VIDEO
COMMENTS
To assign structures to a spectrum: 1. Open a spectral file. ... From the Structure menu, choose Make Spectrum-Structure Assignment. The selected atoms and bonds in the structure are associated with the selected spectral peaks. CambridgeSoft Corporation CambridgeSoft.com Voice: 1 800 315-7300 1 617 588-9300
Not sure how accurate they will be. Draw the structure in Chem3D, then go to "calculations" -> "GAMESS Interface" -> "Predict IR/Raman Spectrum". Select your parameters and hit run. EDIT: I was curious so I ran a really quick ab init. calculation on cyclohexane. Looks alright to a reasonable approximation.
1. Open ChemDraw Ultra 10.0 software. 2. Select "Structure" from the menu bar. (circled in red) 3. Select "Convert Name to Structure" from the pull down menu. (circled in green) 4. The Insert Structure box will pop-up. Enter the name of the molecule you would like to draw and click OK. 5. The molecule structure will appear.
For a typical structure assignment of a small organic molecule (e.g., fewer than ∼10 non-H atoms or up to ∼180 a.m.u. and ∼20 conformers), this protocol can be completed in ∼2 h of active ...
introduced. The primary addition here involves adding a compound structure and making assignments directly to it. a) Use ChemDraw or ChemSketch to copy-and-paste a compound structure into your 1H 1D spectrum; CNTL-C and CNTL-V work fine for this. Note: I (cgf) have been unable to get renumbering in ChemDraw to replicate in MNova,
Features of ChemDraw:Chemical structure to name conversionChemical name to structure conversionNMR spectrum simulation (1H and 13C)Mass spectrum simulationSt...
When finished with the partial structure column, you should be able to link the partial structures together to make an entire structure in the assignment column. An example of a spectrum and its accompanying data table is given below. Here is the spectrum: Here is a data table: Things to note: This student has used two integration columns ...
In ChemDraw Pro, break existing structures across one or more bonds to mimic the fragmentation in a mass spectrometer. In ChemDraw Ultra, assign structures to spectra and calculate NMR shift information. CambridgeSoft Corporation CambridgeSoft.com Voice: 1 800 315-7300 1 617 588-9300
This is a two part experiment: Part 1: You will use the Darling Framework Molecular Models to complete several exercises that explore equivalent structures, equivalent atoms within structures, constitutional isomers, and cis-trans isomers. Your particular assignment will be given to you at lab time. Do not forget to bring the model kit with you!!
Go to "Object" and make sure "fixed length" and "fixed angles" are both checked. 1.2 Copy and paste. In general for one project you should use the same document. You can expand the size of your ChemDraw workspace by changing the document size under "File", "Document Settings". SAVE FREQUENTLY!!!
Find the structure from 1H spectrum. 1H exercise generator. Assign 1H NMR spectra to molecule. 13C NMR. 1H NMR spectra of small molecules. 1H NMR spectra of Boc amino acids. Number of different Hs. 1H NMR integrate and find the structure. 1H number of signals.
Both utilities will provide a molecular formula and calculated molecular weight for peptide/peptoid sequences entered, in addition to an automatically generated ChemDraw structure in .cdxml format (Fig. 2). A spectral assignment for the peptide/peptoid will also be given if m/z values were provided as part of the input.
This demo will simulate 1 H and 13 C NMR spectra, as well as the mass spectrum parent peak (isotopic distribution), of the molecule you draw in the sketcher. Click the Simulate Spectra button to simulate the spectra when you finish drawing your molecule. The spectra are interactive, so you can change their perspectives. For more simulation options, atom assignments and publishing features ...
For a typical structure assignment of a small organic molecule ... simple structures. Namely, because ChemDraw (via the 'Predict ... mental spectrum of 1-trans or 1-cis. One commonly used method
S - Resource - How to draw 3D chemdraw structures.cdx. To draw 3-dimensionalstructures in Chemdraw one can draw the molecule in a 3D viewer and then paste the png here and go over it. The tridimensional effect can be reinforced using bold bonds. This is an example.
A step-by-step guide to drawing chemical structures with ChemDraw. This website uses cookies to improve website functionality and performance, to analyze website traffic, and to provide you with a more personalized experience. ... Tutorial guide to ChemDraw . From Andrew Nortcliffe likes views comments. Related Media. Details; Back. A step-by ...
4 — Short Manual to ChemDraw Drawing Structures When starting the ChemDraw program, a new blank document or a previously used document opens. In addition to the window on which to draw, a vertical palette of tools ("Main Toolbar") on ... To plan a scheme or a drawing of a structure, make a hand-sketched draft. Drawing the Framework of a ...
Both utilities will provide a molecular formula and calculated molecular weight for peptide/peptoid sequences entered, in addition to an automatically generated ChemDraw structure in .cdxml format (Fig. (Fig.2). 2). A spectral assignment for the peptide/peptoid will also be given if m/z values were provided as part of the input.
To draw and search on a structure or substructure, click "Structure search," then click the pencil inside the diagram, then select "Draw or Edit". PubChem. PubChem Structure Search allows the PubChem Compound Database to be to be queried by chemical structure or chemical structure pattern. The PubChem Sketcher allows a query to be drawn ...
ChemBioDraw (commonly called ChemDraw by chemists) is a program designed for drawing molecules and reactions. The software is easy to use, but the more time you spend exploring available options and practices using the program, the easier it will be to properly complete your future lab reports. In the assignment, you will be asked to complete ...
Then follow the menu 'Analysis/Assignment' (or use the shortcut 'A'). Click on an atom on the molecular structure (or a spectrum region) and then release the mouse and drag it to your desired peak. Once your desired peak is highlighted on the spectrum, click on it to assign it. This peak will now be assigned to the atom (which will turn to green).
Structure菜单中有什么命令?. Structure菜单是ChemDraw工具栏中的重要成员之一,熟练掌握的话大大有助于高效绘制化学结构,菜单命令有以下几个方向:. 1、属性管理:AtomProperties、Bond Properties和Bracket Properties可以分别打开原子、化学键以及括号属性设置框,用户 ...