Nucleic Acids Research (NAR) publishes the results of leading edge research into physical, chemical, biochemical and biological aspects of nucleic acids and proteins involved in nucleic acid metabolism and/or interactions. It enables the rapid publication of papers under the following categories: Chemistry and synthetic biology; Computational biology; Gene regulation, chromatin and epigenetics; Genome integrity, repair and replication; Genomics; Molecular biology; Nucleic acid enzymes; RNA and Structural biology. A Survey and Summary section provides a format for brief reviews. The first issue of each year is devoted to biological databases, and an issue in July is devoted to papers describing web-based software resources of value to the biological community.
Some content from Wikipedia , licensed under CC BY-SA

Nucleic Acid Research
- Date 6 hours 12 hours 1 day 3 days all
- Rank Last day 1 week 1 month all
- LiveRank Last day 1 week 1 month all
- Popular Last day 1 week 1 month all
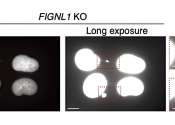
Researchers study the intricacies of homologous recombination and abnormal chromosome bridges
Keeping the genetic information stored in genomic DNA intact during the cell division cycle is crucial for almost all lifeforms. Extensive DNA damage invariably causes various adverse genomic rearrangements, which can lead ...
Cell & Microbiology
May 7, 2024
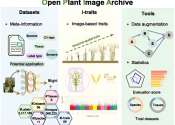
Scientists develop open archive of plant images and related phenotypic traits
Plant images contain a wealth of information that reflects key phenotypic characteristics such as color, shape, growth, and health status of plants. High-throughput plant phenotypic collection technology has been widely applied ...
Molecular & Computational biology
Nov 7, 2023
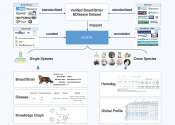
Researchers develop curated and integrated biomarker knowledgebase for animals
Biological markers, commonly referred to markers or biomarkers, serve as quantifiable and measurable indicators of certain biological states in normal and pathogenic processes, as well as potential pharmacologic responses ...
Plants & Animals
Nov 2, 2023
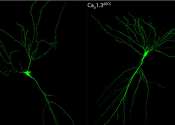
Loss of RNA editing in the Caᵥ1.3 channel to enhance spatial memory comes at a price
In biology, it has been observed that genetic information flows from genomic DNA to messenger RNA and finally, to the protein. The genetic information embedded within the DNA has been touted as the "Language of Life".
Aug 12, 2022

Autonomous nanomachines inspired by nature
Inspired by the way molecules interact in nature, UNSW medical researchers engineer versatile nanoscale machines to enable greater functional range.
Bio & Medicine
Mar 23, 2022
PopHumanVar: Reconstructing the evolutionary past of the human species
A UAB research team has developed PopHumanVar, an application that helps reconstructing the evolutionary past of the human species through the identification of specific genetic mutations that have allowed us to adapt to ...
Feb 2, 2022

High-resolution mapping method for G4 DNA structures developed
While investigating the unusual G quadruplex DNA structure (G4), the Simon Elsässer group has developed a more accurate method for mapping these structures in the genome. G4 CUT&Tag revealed numerous G4s in the human and ...
Biotechnology
Dec 6, 2021
Pathogenic bacteria rendered almost harmless
Pseudomonas aeruginosa is an opportunistic pathogenic bacterium present in many ecological niches, such as plant roots, stagnant water or even the pipes of our homes. Naturally very versatile, it can cause acute and chronic ...
Jun 21, 2021

Solving the puzzle pieces of the mitochondrial ribosome assembly
Researchers at Karolinska Institutet characterize a GTPase involved in ribosome biogenesis in mitochondria. The study, published in Nucleic Acid Research, reveals new details of a complex process that enables mitochondria ...
Dec 7, 2020

Hunt for interesting metabolites with the antiSMASH database
Scientists who hunt for interesting bacterial metabolites using the online tool antiSMASH now have the opportunity to use an antiSMASH database with pre-calculated results of nearly 25,000 bacterial genomes. This database ...
Nov 8, 2018
E-mail newsletter
- Search Menu
- Chemical Biology and Nucleic Acid Chemistry
- Computational Biology
- Critical Reviews and Perspectives
- Data Resources and Analyses
- Gene Regulation, Chromatin and Epigenetics
- Genome Integrity, Repair and Replication
- Methods Online
- Molecular Biology
- Nucleic Acid Enzymes
- RNA and RNA-protein complexes
- Structural Biology
- Synthetic Biology and Bioengineering
- Advance Articles
- Breakthrough Articles
- Special Collections
- Scope and Criteria for Consideration
- Author Guidelines
- Data Deposition Policy
- Database Issue Guidelines
- Web Server Issue Guidelines
- Submission Site
- About Nucleic Acids Research
- Editors & Editorial Board
- Information of Referees
- Self-Archiving Policy
- Dispatch Dates
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Advance articles
Chip-atlas 3.0: a data-mining suite to explore chromosome architecture together with large-scale regulome data.

- View article
- Supplementary data
Orphan nuclear receptors-induced ALT-associated PML bodies are targets for ALT inhibition

Annotation and visualization of parasite, fungi and arthropod genomes with Companion

KinomeMETA: a web platform for kinome-wide polypharmacology profiling with meta-learning

asteRIa enables robust interaction modeling between chromatin modifications and epigenetic readers

Two residues in the DNA binding site of Pif1 helicase are essential for nuclear functions but dispensable for mitochondrial respiratory growth

DNAforge: a design tool for nucleic acid wireframe nanostructures

AIUPred: combining energy estimation with deep learning for the enhanced prediction of protein disorder

TEENA: an integrated web server for transposable element enrichment analysis in various model and non-model organisms

Deep learning for the PSIPRED Protein Analysis Workbench

AlphaFind: discover structure similarity across the proteome in AlphaFold DB

Genome editing outcomes reveal mycobacterial NucS participates in a short-patch repair of DNA mismatches

A-MYB substitutes for B-MYB in activating cell cycle genes and in stimulating proliferation

NF-κB factors cooperate with Su(Hw)/E4F1 to balance Drosophila /human immune responses via modulating dynamic expression of miR-210

Evaluating the oral delivery of GalNAc-conjugated siRNAs in rodents and non-human primates

DORA: an interactive map for the visualization and analysis of ancient human DNA and associated data

Riboswitch and small RNAs modulate btuB translation initiation in Escherichia coli and trigger distinct mRNA regulatory mechanisms

SEMA 2.0: web-platform for B-cell conformational epitopes prediction using artificial intelligence

The mutation R107Q alters mtSSB ssDNA compaction ability and binding dynamics

Single-molecule imaging reveals a direct role of CTCF’s zinc fingers in SA interaction and cluster-dependent RNA recruitment

Structural basis for double-stranded RNA recognition by SID1

Stress-induced nucleoid remodeling in Deinococcus radiodurans is associated with major changes in Heat Unstable (HU) protein dynamics

Aggrescan4D: structure-informed analysis of pH-dependent protein aggregation

PROSCA: an online platform for humanized scaffold mining facilitating rational protein engineering

Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand

Kinetic pathway of HIV-1 TAR cotranscriptional folding

LncRNAway: a web-based sgRNA design tool for precise and effective suppression of long noncoding RNAs

SimRNAweb v2.0: a web server for RNA folding simulations and 3D structure modeling, with optional restraints and enhanced analysis of folding trajectories

GPSFun: geometry-aware protein sequence function predictions with language models

Visualization of oxidized guanine nucleotides accumulation in living cells with split MutT

Drawing human pedigree charts with DrawPed

AcrIIA28 is a metalloprotein that specifically inhibits targeted-DNA loading to SpyCas9 by binding to the REC3 domain

A programmable dual-targeting siRNA scaffold supports potent two-gene modulation in the central nervous system

Massively parallel dissection of RNA in RNA–protein interactions in vivo

Constraints on the emergence of RNA through non-templated primer extension with mixtures of potentially prebiotic nucleotides

Reduction of ZFX levels decreases histone H4 acetylation and increases Pol2 pausing at target promoters

EXO1 and DNA2-mediated ssDNA gap expansion is essential for ATR activation and to maintain viability in BRCA1-deficient cells

Increased PTCHD4 expression via m 6 A modification of PTCHD4 mRNA promotes senescent cell survival

Genome-wide analysis of mRNA decay in Arabidopsis shoot and root reveals the importance of co-translational mRNA decay in the general mRNA turnover

Comprehensive translational profiling and STE AI uncover rapid control of protein biosynthesis during cell stress

Skipping events impose repeated binding attempts: profound kinetic implications of protein–DNA conformational changes

Mibianto: ultra-efficient online microbiome analysis through k -mer based metagenomics

Transcription factor shapes chromosomal conformation and regulates gene expression in bacterial adaptation

Identification and in vitro characterization of UDP-GlcNAc-RNA cap-modifying and decapping enzymes

Prokaryotic Argonaute nuclease cooperates with co-encoded RNase to acquire guide RNAs and target invader DNA

Fluoropyrimidines trigger decay of hypomodified tRNA in yeast

GPS-SUMO 2.0: an updated online service for the prediction of SUMOylation sites and SUMO-interacting motifs

Arrayed in vivo barcoding for multiplexed sequence verification of plasmid DNA and demultiplexing of pooled libraries

SCTC: inference of developmental potential from single-cell transcriptional complexity

Structural snapshots of phenuivirus cap-snatching and transcription

mtDNA-Server 2: advancing mitochondrial DNA analysis through highly parallelized data processing and interactive analytics

A novel regulatory interplay between atypical B 12 riboswitches and uORF translation in Mycobacterium tuberculosis

SHAPEwarp-web: sequence-agnostic search for structurally homologous RNA regions across databases of chemical probing data

IsoVis – a webserver for visualization and annotation of alternative RNA isoforms

LipidSig 2.0: integrating lipid characteristic insights into advanced lipidomics data analysis

A bisulfite-assisted and ligation-based qPCR amplification technology for locus-specific pseudouridine detection at base resolution

Imputation Server PGS: an automated approach to calculate polygenic risk scores on imputation servers

UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2

PRMT5-mediated arginine methylation of FXR1 is essential for RNA binding in cancer cells

Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment

Correction to ‘The Uba4 domain interplay is mediated via a thioester that is critical for tRNA thiolation through Urm1 thiocarboxylation’
Correction to ‘global hfq-mediated rna interactome of nitrogen starved escherichia coli uncovers a conserved post-transcriptional regulatory axis required for optimal growth recovery’, correction to ‘rnase e searches for cleavage sites in rna by linear diffusion: direct evidence from single-molecule fret’, swissdock 2024: major enhancements for small-molecule docking with attracting cavities and autodock vina.

RING 4.0: faster residue interaction networks with novel interaction types across over 35,000 different chemical structures

ChemoDOTS: a web server to design chemistry-driven focused libraries

BGCFlow: systematic pangenome workflow for the analysis of biosynthetic gene clusters across large genomic datasets

The RBPome of influenza A virus NP-mRNA reveals a role for TDP-43 in viral replication

Systematic identification of cargo-mobilizing genetic elements reveals new dimensions of eukaryotic diversity

The uS10c-BPG2 module mediates ribosomal RNA processing in chloroplast nucleoids

SynDesign: web-based prime editing guide RNA design and evaluation tool for saturation genome editing

DNA-PK participates in pre-rRNA biogenesis independent of DNA double-strand break repair

Functional redundancy in tRNA dihydrouridylation

Senescence of human pancreatic beta cells enhances functional maturation through chromatin reorganization and promotes interferon responsiveness

Short 2′- O -methyl/LNA oligomers as highly-selective inhibitors of miRNA production in vitro and in vivo

Controlling genome topology with sequences that trigger post-replication gap formation during replisome passage: the E. coli RRS elements

iM-Seeker: a webserver for DNA i-motifs prediction and scoring via automated machine learning

SpatialSPM: statistical parametric mapping for the comparison of gene expression pattern images in multiple spatial transcriptomic datasets

Simultaneous profiling of chromatin accessibility and DNA methylation in complete plant genomes using long-read sequencing

A nematode-specific ribonucleoprotein complex mediates interactions between the major nematode spliced leader snRNP and its target pre-mRNAs

CARM1 hypermethylates the NuRD chromatin remodeling complex to promote cell cycle gene expression and breast cancer development

Correction to ‘The levels of p53 govern the hierarchy of DNA damage tolerance pathway usage’
Correction to ‘human cdk18 promotes replication stress signaling and genome stability’, correction to ‘pgs-depot: a comprehensive resource for polygenic scores constructed by summary statistics based methods’, multivalent dnazyme agents for cleaving folded rna.

TTSBBC: triplex target site biomarkers and barcodes in cancer

The Damietta Server: a comprehensive protein design toolkit

Molecular mechanism for target RNA recognition and cleavage of Cas13h

Precise editing of pathogenic nucleotide repeat expansions in iPSCs using paired prime editor

Pred-O3, a web server to predict molecules, olfactory receptors and odor relationships


Structural and biochemical characterization of the mitomycin C repair exonuclease MrfB

Epigenetic control of adaptive or homeostatic splicing during interval-training activities

The ABCF proteins in Escherichia coli individually cope with ‘hard-to-translate’ nascent peptide sequences

Systematic interrogation of CRISPR antimicrobials in Klebsiella pneumoniae reveals nuclease-, guide- and strain-dependent features influencing antimicrobial activity

Towards parsimonious generative modeling of RNA families

Expression of Concern on ‘MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation’
Admetsar3.0: a comprehensive platform for exploration, prediction and optimization of chemical admet properties.

Comprehensive genomic features indicative for Notch responsiveness

High-density generation of spatial transcriptomics with STAGE

Email alerts
- Editorial Board
Affiliations
- Online ISSN 1362-4962
- Print ISSN 0305-1048
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
The 2021 Nucleic Acids Research database issue and the online molecular biology database collection
Affiliations.
- 1 Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Crown Street, Liverpool L69 7ZB, UK.
- 2 Institut Curie, 25 rue d'Ulm, 75005 Paris, France.
- PMID: 33396976
- PMCID: PMC7778882
- DOI: 10.1093/nar/gkaa1216
The 2021 Nucleic Acids Research database Issue contains 189 papers spanning a wide range of biological fields and investigation. It includes 89 papers reporting on new databases and 90 covering recent changes to resources previously published in the Issue. A further ten are updates on databases most recently published elsewhere. Seven new databases focus on COVID-19 and SARS-CoV-2 and many others offer resources for studying the virus. Major returning nucleic acid databases include NONCODE, Rfam and RNAcentral. Protein family and domain databases include COG, Pfam, SMART and Panther. Protein structures are covered by RCSB PDB and dispersed proteins by PED and MobiDB. In metabolism and signalling, STRING, KEGG and WikiPathways are featured, along with returning KLIFS and new DKK and KinaseMD, all focused on kinases. IMG/M and IMG/VR update in the microbial and viral genome resources section, while human and model organism genomics resources include Flybase, Ensembl and UCSC Genome Browser. Cancer studies are covered by updates from canSAR and PINA, as well as newcomers CNCdatabase and Oncovar for cancer drivers. Plant comparative genomics is catered for by updates from Gramene and GreenPhylDB. The entire Database Issue is freely available online on the Nucleic Acids Research website (https://academic.oup.com/nar). The NAR online Molecular Biology Database Collection has been substantially updated, revisiting nearly 1000 entries, adding 90 new resources and eliminating 86 obsolete databases, bringing the current total to 1641 databases. It is available at https://www.oxfordjournals.org/nar/database/c/.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Publication types
- Introductory Journal Article
- Research Support, Non-U.S. Gov't
- COVID-19 / epidemiology
- COVID-19 / prevention & control
- COVID-19 / virology
- Computational Biology / methods
- Databases, Nucleic Acid*
- Genomics / methods
- Molecular Biology / methods
- Molecular Biology / standards
- Molecular Biology / statistics & numerical data*
- Nucleic Acids*
- Periodicals as Topic / standards
- Periodicals as Topic / statistics & numerical data*
- Research / standards
- Research / statistics & numerical data*
- SARS-CoV-2 / genetics*
- SARS-CoV-2 / physiology
- Nucleic Acids
- Directories
Project Info
Synthesis and characterization of polyplexes formed from cationic copolymers and nucleic acids., project goals and description:, more information:, primary contacts:, student preparation, qualifications, time commitment (hrs/wk), skills/techniques gained, mentoring plan, preferred student status.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Nucleic acids articles within Nature Communications
Article 09 April 2024 | Open Access
Expedient production of site specifically nucleobase-labelled or hypermodified RNA with engineered thermophilic DNA polymerases
A general method for enzymatic synthesis of base-modified RNA was developed using engineered thermostable DNA polymerases enabling introduction of site-specific modifications or synthesis of hypermodified RNA not accessible by in vitro transcription.
- Mária Brunderová
- , Vojtěch Havlíček
- & Michal Hocek
Article 21 March 2024 | Open Access
Late-stage guanine C8–H alkylation of nucleosides, nucleotides, and oligonucleotides via photo-mediated Minisci reaction
Chemically modified nucleobases and oligonucleotides are essential in several fields but introducing functional groups into nucleobases requires laborious chemical synthesis. Here, the authors report site-selective alkylation at the C8-position of guanines in guanosine, GMP, GDP, and GTP, as well as late-stage alkylation of RNA/DNA oligonucleotides through photomediated Minisci reaction.
- Ruoqian Xie
- & Gang Chen
Article 15 February 2024 | Open Access
Trabectedin derails transcription-coupled nucleotide excision repair to induce DNA breaks in highly transcribed genes
The antitumor drug trabectedin is more toxic to DNA-repair-proficient cells. Here the authors show that this is caused by persistent DNA breaks induced from an abortive repair reaction and develop “TRABI-Seq” to map the breaks to transcribed regions of the genome. Trabectedin may thus be used as a diagnostic and therapeutic in precision oncology.
- , Vakil Takhaveev
- & Orlando D. Schärer
Article 12 February 2024 | Open Access
Liquid crystalline inverted lipid phases encapsulating siRNA enhance lipid nanoparticle mediated transfection
The authors display the bottom-up design, assembly, and in-depth characterization of defined lipid-RNA structures in the core of lipid nanoparticles. The inverted structures are thermostable and provide better transfection over lamellar structures.
- Roy Pattipeiluhu
- & Thomas H. Sharp
Article 15 November 2023 | Open Access
Profiling stress-triggered RNA condensation with photocatalytic proximity labeling
Stress granules (SGs) are highly dynamic cytoplasmic membraneless organelles that assemble when cells are challenged by stress. Herein, the authors apply a proximity-dependent RNA labeling method, CAP-seq, to comprehensively investigate the content of SG-proximal transcriptome and the dynamic change in SG-proximal transcriptome along the time course of granule assembly and disassembly processes in live mammalian cells.
- & Peng Zou
Article 26 October 2023 | Open Access
Enzymatic synthesis and nanopore sequencing of 12-letter supernumerary DNA
Unnatural base pairing xenonucleic acids (XNAs) can be used to expand life’s alphabet beyond ATGC. Here, authors show strategies for enzymatic synthesis and next-generation nanopore sequencing of XNA base pairs for reading and writing 12-letter DNA (ATGCBSPZXKJV).
- Hinako Kawabe
- , Christopher A. Thomas
- & Jorge A. Marchand
Article 17 August 2023 | Open Access
Extensive breaking of genetic code degeneracy with non-canonical amino acids
Genetic code expansion is limited by the degeneracy of the 61 sense codons which encode for only 20 amino acids. Here, the authors show that by combining hyperaccurate ribosomes and in vitro transcribed tRNAs, dramatic and extensive breaking of sense codon degeneracy can be achieved.
- Clinton A. L. McFeely
- , Bipasana Shakya
- & Matthew C. T. Hartman
Article 13 July 2023 | Open Access
Hidden modes of DNA binding by human nuclear receptors
Nuclear receptors (NR) are drug-responsive master regulators. Here, authors map DNA binding profiles of all human NRs. Their MinSeq Find algorithm identifies masked NR binding sites in genomes and maps ~10% of orphan SNPs linked to numerous diseases.
- Devesh Bhimsaria
- , José A. Rodríguez-Martínez
- & Aseem Z. Ansari
Article 27 April 2023 | Open Access
Chemical evolution of an autonomous DNAzyme with allele-specific gene silencing activity
Low activity currently prevents the wider use of DNA enzymes (DNAzymes). Here the authors report the chemical evolution of a DNAzyme with high catalytic activity under near physiological conditions: the enzyme achieves ~65 turnovers in 30 minutes.
- , Turnee N. Malik
- & John C. Chaput
Article 13 January 2023 | Open Access
Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase
T7 RNA polymerase (RNAP) is widely used for synthesizing RNA molecules with synthetic modifications and unnatural base pairs (UBPs). Here, authors show the structural basis of how UBPs are recognized as template and substrate, providing mechanistic insights into UBP transcription by T7 RNAP.
- , Michiko Kimoto
- & Dong Wang
Article 10 January 2023 | Open Access
Additive-controlled asymmetric iodocyclization enables enantioselective access to both α- and β-nucleosides
Nucleosides and their analogs are pharmacologically important molecules. Here, the authors report an additive-controlled stereodivergent iodocyclization method for the selective synthesis of α- or β-nucleosides.
- & Fener Chen
Article 29 November 2022 | Open Access
Shedding light on the base-pair opening dynamics of nucleic acids in living human cells
Base-pair opening is important for nucleic acids to exert biological functions, but studying its dynamics inside living cells is challenging. Here, the authors determine the base-pair opening kinetics of hairpin and G-quadruplex structures inside living human cells by the in-cell NMR technique, and demonstrate a difference in dynamics of nucleic acids between in-cell and in vitro conditions.
- Yudai Yamaoki
- , Takashi Nagata
- & Masato Katahira
Article 16 November 2022 | Open Access
XNAzymes targeting the SARS-CoV-2 genome inhibit viral infection
RNA viruses have been responsible for large-scale epidemics and pandemics throughout the last few centuries. Here, the authors show the design, synthesis and screening of artificial RNA endonuclease XNAzymes capable of cleaving genomic SARS-CoV-2 RNA and self-assembling into enzymatic nanostructures inhibiting cellular viral replication.
- Pehuén Pereyra Gerber
- , Maria J. Donde
- & Alexander I. Taylor
Article 02 November 2022 | Open Access
HIF1α-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex
Using a composite bioinformatics approach, the DNA:DNA:RNA triplex-forming lncRNAs HIF1α-AS1 was identified in human endothelial cells which recruits an epigenetic silencing complex to limit expression of triplex target genes.
- Matthias S. Leisegang
- , Jasleen Kaur Bains
- & Ralf P. Brandes
Article 29 October 2022 | Open Access
Microfluidic space coding for multiplexed nucleic acid detection via CRISPR-Cas12a and recombinase polymerase amplification
Fast, low-cost and multiplexed nucleic acid detection is challenging. Here the authors report a strategy that couples microfluidic space coding, CRISPRCas12a, and multiplex RPA for the rapid detection of up to 30 targets with only one fluorescent probe.
- , Dongjuan Chen
- & Maili Liu
Article 08 October 2022 | Open Access
Controllable DNA hybridization by host–guest complexation-mediated ligand invasion
Direct dissociation of nucleic acid duplex structures without heating or specific binding proteins is challenging. Here the authors use the cucurbit[7]uril-based host–guest system to construct a ligand-invasion pathway for controllable DNA hybridisation.
- , Liang-Liang Wang
- & Liang Xu
Article 26 August 2022 | Open Access
Efficient DNA fluorescence labeling via base excision trapping
Methods for fluorescently labelling DNAs are expensive and labour-intensive. Here the authors report an in situ DNA labelling strategy for oligonucleotides as well as dsDNA that makes use of aldehyde-reactive rotor dyes to trap AP sites resulting from excision of deaminated DNA bases.
- Yong Woong Jun
- , Emily M. Harcourt
- & Eric T. Kool
Article 30 June 2022 | Open Access
Simple synthesis of massively parallel RNA microarrays via enzymatic conversion from DNA microarrays
RNA microarrays have many potential applications, but are difficult to produce. Here, the AUs present a method for converting commercial, customizable DNA microarrays into RNA microarrays using an accessible three-step process involving primer photocrosslinking, extension, and template degradation.
- Erika Schaudy
- , Kathrin Hölz
- & Mark M. Somoza
Article 09 May 2022 | Open Access
Targeting double-strand break indel byproducts with secondary guide RNAs improves Cas9 HDR-mediated genome editing efficiencies
Programmable double-strand DNA breaks (DSBs) can be harnessed for precision genome editing through manipulation of the homology-directed repair (HDR) pathway. Here the authors report the development of the double tap - double tap implements secondary gRNAs which target Cas9 to common indel sequences and provides a second chance at HDR.
- Zsolt Bodai
- , Alena L. Bishop
- & Alexis C. Komor
Article 28 March 2022 | Open Access
Structural basis of R-loop recognition by the S9.6 monoclonal antibody
The S9.6 monoclonal antibody is widely used to map R-loops genome wide. Here, Bou-Nader et al., define the nucleic acid-binding specificity of S9.6 and report its crystal structures free and bound to a hybrid, which reveal the asymmetric recognition of the RNA and DNA strands and its A-form conformation.
- Charles Bou-Nader
- , Ankur Bothra
- & Jinwei Zhang
Article 15 November 2021 | Open Access
Gene editing with CRISPR-Cas12a guides possessing ribose-modified pseudoknot handles
Development of Cas12a for human therapeutics and diagnostics may significantly benefit from, or even require, chemical modification of its guide RNA. Here the authors show that the noncanonical 5′ pseudoknot structure of the AsCas12a crRNA guide can be heavily modified and still retain very high editing activity in cells and trans cleavage activity in vitro.
- Eman A. Ageely
- , Ramadevi Chilamkurthy
- & Keith T. Gagnon
Article 06 October 2021 | Open Access
A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites
The small modified nucleotide PreQ1 binds to the PreQ1 riboswitch and regulates gene expression by inducing RNA conformation change. Here the authors design and characterize a specific preQ1-derived probe by x-ray crystallography, mass spectral analysis and transcriptome-wide using Chem-CLIP.
- Sumirtha Balaratnam
- , Curran Rhodes
- & John S. Schneekloth Jr
Article 24 August 2021 | Open Access
A non-enzymatic, isothermal strand displacement and amplification assay for rapid detection of SARS-CoV-2 RNA
The reliance on enzymes in SARS-CoV-2 RNA detection imposes limits on transport and storage conditions. Here the authors use non-enzymatic isothermal amplification to detect RNA with no need for reverse transcription.
- Mohsen Mohammadniaei
- , Ming Zhang
- & Yi Sun
Article 02 August 2021 | Open Access
A kinetically controlled platform for ligand-oligonucleotide transduction
Ligand-oligonucleotide interactions can integrate both small molecules and proteins into nucleic acid-based circuits. Here the authors design ligand-aptamer complexes to control strand-displacement reactions for versatile ligand transduction.
- Qiu-Long Zhang
Article 20 July 2021 | Open Access
Fully automated fast-flow synthesis of antisense phosphorodiamidate morpholino oligomers
PMOs (phosphorodiamidate morpholino oligomers) have huge potential for antisense therapy but complex and slow synthesis limits application. Here, the authors report the development of automated flow synthesis methods which reduce nucleobase coupling times from hours to minutes removing human errors and allow for high-throughput production.
- , Alex J. Callahan
- & Bradley L. Pentelute
Article 05 February 2021 | Open Access
Nonenzymatic polymerase-like template-directed synthesis of acyclic l -threoninol nucleic acid
A world preceding the prebiotic RNA-world may have been based on xeno nucleic acids (XNAs), but their replication likely did not require enzymes. Here, the authors demonstrate template-directed non-enzymatic synthesis of an XNA, acyclic l -threoninol nucleic acid, via chemical ligation mediated by N-cyanoimidazole, and achieve a pseudo-primer extension of this XNA with all four nucleobases.
- Keiji Murayama
- , Hikari Okita
- & Hiroyuki Asanuma
Article 24 September 2020 | Open Access
Optoribogenetic control of regulatory RNA molecules
Short hairpin RNAs can be used to modulate and regulate gene expression. Here the authors generate chimeric RNAs that interact with the photoreceptor PAL, allowing for optoribogenetic control of cell physiology.
- Sebastian Pilsl
- , Charles Morgan
- & Günter Mayer
Article 03 January 2020 | Open Access
Conditional control of RNA-guided nucleic acid cleavage and gene editing
Constituitively active CRISPR systems have the risk of adverse off-target effects. Here the authors use chemical masking and activation of gRNA to control activity.
- Shao-Ru Wang
- , Ling-Yu Wu
- & Xiang Zhou
Article 16 December 2019 | Open Access
SAM-VI riboswitch structure and signature for ligand discrimination
Riboswitches are conserved RNA domains located in the non-coding region of mRNA that recognize cellular metabolites and, in turn, regulate gene expression. Here the authors report the structure of the recently identified SAM-VI riboswitch and provide insight into its mechanism of ligand discrimination.
- , Catherina Gasser
- & Aiming Ren
Article 29 July 2019 | Open Access
Time-resolved NMR monitoring of tRNA maturation
Transfer RNA (tRNA) is regulated by RNA modifications. Here the authors employ time-resolved NMR to monitor modifications of yeast tRNA Phe in cellular extracts, revealing a sequential order and cross-talk between modifications.
- Pierre Barraud
- , Alexandre Gato
- & Carine Tisné
Article 08 April 2019 | Open Access
An artificial triazole backbone linkage provides a split-and-click strategy to bioactive chemically modified CRISPR sgRNA
For CRISPR-Cas9 genome editing, Cas9 protein is guided to its target by single guide (sg) RNA. Here, the authors synthesised sgRNAs via convergent ‘click’ ligation of variable 20-mer RNAs that target the genome and a Cas9-binding 79-mer chimeric RNA/2´-OMe RNA of fixed sequence in a single tube.
- Lapatrada Taemaitree
- , Arun Shivalingam
- & Tom Brown
Article 04 March 2019 | Open Access
Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9
Amphihelical antimicrobial peptides (AMPs) are bactericidal host defense factors, but their function as immunomodulators is emerging. Here the authors show that several AMPs organize DNA into periodic nanocrystals by self-assembling into superhelical protofibril scaffolds, which potentiates DNA sensing by TLR9.
- Ernest Y. Lee
- , Changsheng Zhang
- & Gerard C. L. Wong
Article 30 January 2019 | Open Access
Template-directed RNA polymerization and enhanced ribozyme catalysis inside membraneless compartments formed by coacervates
Membraneless compartments have been theorized to be prebiotic micro-compartments as they spontaneously encapsulate RNA and proteins. Here, the authors report membraneless compartments can enhance RNA chemistries, affecting template directed RNA polymerization and stimulating nucleic acid enzymes.
- Raghav R. Poudyal
- , Rebecca M. Guth-Metzler
- & Philip C. Bevilacqua
Comment 12 December 2018 | Open Access
Non-canonical nucleosides and chemistry of the emergence of life
- Sidney Becker
- , Christina Schneider
- & Thomas Carell
Prebiotic plausibility and networks of paradox-resolving independent models
- Steven A. Benner
Life as a guide to prebiotic nucleotide synthesis
- Stuart A. Harrison
- & Nick Lane
Prebiotic chemistry and human intervention
- Clemens Richert
Searching for lost nucleotides of the pre-RNA World with a self-refining model of early Earth
- Nicholas V. Hud
Prebiotic nucleic acids need space to grow
- Daniel Whitaker
- & Matthew W. Powner
Experimentally investigating the origin of DNA/RNA on early Earth
- Ramanarayanan Krishnamurthy
Article 19 November 2018 | Open Access
Translation of non-standard codon nucleotides reveals minimal requirements for codon-anticodon interactions
The recognition of the mRNA codon by the tRNA anticodon is crucial for protein synthesis. Here the authors introduce non-standard nucleotides in bacterial and eukaryotic mRNA to reveal the minimal hydrogen bond requirement of codon-anticodon interaction for efficient and accurate translation.
- Thomas Philipp Hoernes
- , Klaus Faserl
- & Matthias David Erlacher
Article 12 October 2018 | Open Access
Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex
Targeting noncoding nucleic acids with small molecules represents an important and significant challenge in chemical biology and drug discovery. Here the authors characterize DC-34, a small molecule that exhibits selective binding to specific G4 structures, and provide a structural basis for its selectivity
- David R. Calabrese
- , Xiang Chen
- & John S. Schneekloth Jr.
Article 11 June 2018 | Open Access
An anionic phthalocyanine decreases NRAS expression by breaking down its RNA G-quadruplex
Hyperactivity of the gene NRAS contributes to the proliferation and metastatic nature of many types of cancer cells. Here, the authors show that NRAS can be controlled by an anionic phthalocyanine coordinating Zn 2+ in combination with photo-irradiation.
- Keiko Kawauchi
- , Wataru Sugimoto
- & Daisuke Miyoshi
Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines
Large RNAs and ribonucleoprotein complexes have shown potential as novel therapeutic agents, but their targeted delivery to cells is still challenging. Here the authors present a modular aptamer nanostructure for intracellular delivery of RNAs up to 250 nucleotides to cancer cells.
- David Porciani
- , Leah N. Cardwell
- & Donald H. Burke
Article 24 January 2018 | Open Access
Comprehensive profiling of the ligand binding landscapes of duplexed aptamer families reveals widespread induced fit
Duplexed aptamers are a common biosensor format; however, how complementary strand sequence, length, and position modulate ligand binding is not well understood. Here, the authors introduce ACE-Scan to comprehensively map binding landscapes, uncovering hotspots of enhanced binding by induced fit.
- Jeffrey D. Munzar
- & David Juncker
Article 27 November 2017 | Open Access
Structural basis for TNA synthesis by an engineered TNA polymerase
The laboratory-evolved polymerase Kod-RI catalyzes α-L-threose nucleic acid (TNA) synthesis. Here, the authors present Kod-RI crystal structures that give insights into how TNA triphosphates are selected and extended in a template-dependent manner, which will help to engineer improved TNA polymerases for synthetic genetics applications.
- Nicholas Chim
- , Changhua Shi
Article 14 July 2017 | Open Access
Target guided synthesis using DNA nano-templates for selectively assembling a G-quadruplex binding c-MYC inhibitor
Identification of inhibitors can be accelerated by using the target as a template for ligand formation. Here the authors show that DNA-functionalised magnetic nanoparticles guide templating of G-quadruplex binding c-MYC inhibitors from an array of building blocks, and can be isolated by magnetic decanting.
- Deepanjan Panda
- , Puja Saha
- & Jyotirmayee Dash
Article 04 February 2016 | Open Access
Activating frataxin expression by repeat-targeted nucleic acids
Expansion of the trinucleotide GAA within an intronic FXN RNA can cause Friedreich's Ataxia (FRDA), an incurable genetic disorder. Here, the authors show that anti-GAA duplex RNAs or single-stranded locked nucleic acids increases FXN protein expression in patient-derived cells to levels similar to wild-type cells.
- , Masayuki Matsui
- & David R. Corey
Article 22 April 2015 | Open Access
Ultrasensitive visual read-out of nucleic acids using electrocatalytic fluid displacement
Point-of-care analytical devices are of interest for diagnostic applications where larger scale laboratory instruments are not feasible or available. Here, the authors present a direct read-out colorimetric sensor which uses catalytic gas production to visualize picomolar concentrations of DNA.
- Justin D. Besant
- , Jagotamoy Das
- & Shana O. Kelley
Article 23 August 2011 | Open Access
Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids
Rotational motion and torsional strain affects DNA replication, transcription and repair. Lipfert et al . have developed a new technique that uses freely orbiting magnetic tweezers to measure equilibrium fluctuations and determine the twist of tethered nucleic acid molecules.
- Jan Lipfert
- , Matthew Wiggin
- & Nynke H. Dekker
Browse broader subjects
- Chemical biology
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Global Nucleic Acid Sample Preparation Industry Research 2023-2033: Emergence of Advanced Stabilization Products, Regulated vs. Multimodal Analysis, Exploring New Applications - ResearchAndMarkets.com
The "Nucleic Acid Sample Preparation Market - A Global and Regional Analysis: Focus on Technology, Application, Product, End User, and Country - Analysis and Forecast, 2023-2033" report has been added to ResearchAndMarkets.com's offering.
The global nucleic acid sample preparation market is projected to reach $5,615.9 million by 2033 from $2,922.8 million in 2023, growing at a CAGR of 6.75% during the forecast period 2023-2033
The base year considered for the calculation of the nucleic acid sample preparation market size is 2022. A historical year analysis has been done for the period FY2020-FY2021. The nucleic acid sample preparation market size has been estimated for FY2022 and projected for the period FY2023-FY2033.
The key factors driving the growth of the global nucleic acid sample preparation market include the growth in the number of genetic tests globally and an increase in demand for reliable next-generation sequencing (NGS) platforms in clinical laboratories. The rapid growth of genomic research has resulted in an increasing need for the storage of biologic samples, including DNA and RNA. Stabilization products enable researchers to transport and store biological samples for long periods with complete and rapid sample recovery at affordable costs.
These products are used to retain biological materials at room temperature without degradation. These stabilization products ensure that the nucleic material, which has been carefully purified, remains safe and ready for further downstream applications, such as ensuring accurate gene expression analysis through qPCR or QRT-PCR. Research studies suggest that cell lysates created using lysis and stabilization buffers observed no shift in Ct value versus fresh samples, indicating that the nucleic acid was stable and intact while stored in these stabilization solutions.
In order to cross-examine biological samples for changes in DNA and RNA efficiently, there have been considerable advances in NGS chemistries, platforms, and bioinformatics pipelines. Currently, the regulated approach requires the use of two separate workflows for library preparation from separate DNA and RNA isolates. The regulated analysis approach presents certain limitations, such as the large amounts of sample required for generating sufficient nucleic acid for multiple workflows, inefficient use of resources, and long turn-around times.
China dominated the Asia-Pacific nucleic acid sample preparation market in 2022. China has maintained itself as an attractive market for life sciences solution providers capable of tapping the strong demand for nucleic acid extraction products. Several companies, including Qiagen, Thermo Fisher Scientific, BioTeke, Promega, F. Hoffmann-La Roche AG, and Takara Bio, are the leading suppliers of nucleic acid extraction consumables in the country. The Chinese government recognizes the potential of nucleic acid sample preparation and is actively promoting its development and manufacturing.
Revenues of the companies have been referenced from their annual reports for FY2020-FY2022. For private companies, revenues have been estimated based on factors such as inputs obtained from primary research, funding history, product approval status, market collaborations, and operational history.
Market Segmentation Highlights
- Based on product, the nucleic acid sample preparation market has been led by consumables, which held a 67.83% share in 2022.
- Based on consumables market by type, the nucleic acid sample preparation market has been led by Kits, which held a 87.65% share in 2022.
- Based on nucleic acid sample preparation consumables market Kits by Type, the nucleic acid sample preparation market has been led by DNA/RNA Sample Isolation/Extraxtion/Purification, which held an 83.93% share in 2022.
- Based on consumables market by analyte, the nucleic acid sample preparation market has been led by DNA, which held a 64.57% share in 2022.
- Based on consumable-based technology, the nucleic acid sample preparation consumable market has been led by silica-based technology, which held a 48.58% share in 2022.
- Based on instrument-based technology, the nucleic acid sample preparation instrument market has been led by magnetic bead-based, which held a 58.72% share in 2022.
- Based on application, the nucleic acid sample preparation market has been led by qPCR, which held a 31.87% share in 2022.
- Based on end user, the nucleic acid sample preparation market has been led by academic research institutes and laboratories, which held a 45.95% share in 2022.
Key Attributes:
Key Topics Covered:
Market Drivers
- Growing Number of Genetic Tests
- Increasing Demand for Reliable Next-Generation Sequencing (NGS) Platforms in Clinical Laboratories
- Increasing Prevalence of Infectious Diseases and Cancer Cases
- Rise in the Field of Microbial Sequencing
- Increase in Awareness and Acceptance of Personalized Medicines on a Global Level
- Decreasing Cost of Sequencing
Market Restraints
- Genomic Data Protection
- High Cost of Automated Instruments
- Rigid Regulatory Standards
Market Opportunities
- Development and Utilization of More Biobanks in Healthcare Segment
- Technological Advancements in DNA/RNA Sample Preparation Processes
- Growth in Emerging Nations
- Massive Scope for Adoption of NGS-based Genetic Tests in Emerging Markets
Key Developments
- Product Launches and Approvals
- Collaborations and Agreements
- Acquisitions
- Business Expansions
Industry Trends
- Emergence of Advanced Stabilization Products
- Regulated vs. Multimodal Analysis
- Exploring New Applications
- Other Key Trends
Definition by Products
- Instruments
- Column-Based Instruments
- Bead-Based Instruments
- Low-Throughput Instruments
- Medium-Throughput Instruments
- High-Throughput Instruments
Nucleic Acid Sample Preparation Automation Applications
- Genomic/Plasmid DNA Extraction
- Cell-Free DNA (cfDNA) Extraction
- Viral RNA Extraction
- Cell-Free RNA Extraction
Company Profiles: Company Overview, Role in the Global Nucleic Acid Sample Preparation Market, Financials, Analyst Perspective
- Agilent Technologies, Inc.
- Autogen, Inc.
- Bio-Rad Laboratories, Inc.
- F. Hoffmann-La Roche AG
- Macherey-Nagel GmbH & Co KG
- New England Biolabs, Inc.
- Norgen Biotek Corp.
- Omega Bio-tek, Inc.
- Revvity, Inc. (PerkinElmer, Inc.)
- Promega Corporation
- QIAGEN N.V.
- Sage Science, Inc.
- Tecan Group
- Thermo Fisher Scientific Inc.
- Zymo Research
For more information about this report visit https://www.researchandmarkets.com/r/m12mu1
About ResearchAndMarkets.com
ResearchAndMarkets.com is the world's leading source for international market research reports and market data. We provide you with the latest data on international and regional markets, key industries, the top companies, new products and the latest trends.
View source version on businesswire.com: https://www.businesswire.com/news/home/20240516396931/en/
ResearchAndMarkets.com Laura Wood, Senior Press Manager [email protected] For E.S.T Office Hours Call 1-917-300-0470 For U.S./ CAN Toll Free Call 1-800-526-8630 For GMT Office Hours Call +353-1-416-8900
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Nucleic Acids Res
- Search Menu
- Chemical Biology and Nucleic Acid Chemistry
- Computational Biology
- Critical Reviews and Perspectives
- Data Resources and Analyses
- Gene Regulation, Chromatin and Epigenetics
- Genome Integrity, Repair and Replication
- Methods Online
- Molecular Biology
- Nucleic Acid Enzymes
- RNA and RNA-protein complexes
- Structural Biology
- Synthetic Biology and Bioengineering
- Advance Articles
- Breakthrough Articles
- Special Collections
- Scope and Criteria for Consideration
- Author Guidelines
- Data Deposition Policy
- Database Issue Guidelines
- Web Server Issue Guidelines
- Submission Site
- About Nucleic Acids Research
- Editors & Editorial Board
- Information of Referees
- Self-Archiving Policy
- Dispatch Dates
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Article Contents
Introduction, materials and methods, data availability, supplementary data, acknowledgements, prmt5-mediated arginine methylation of fxr1 is essential for rna binding in cancer cells.
The first two authors should be regarded as Joint First Authors.
- Article contents
- Figures & tables
- Supplementary Data
Anitha Vijayakumar, Mrinmoyee Majumder, Shasha Yin, Charles Brobbey, Joseph Karam, Breege Howley, Philip H Howe, Stefano Berto, Lalima K Madan, Wenjian Gan, Viswanathan Palanisamy, PRMT5-mediated arginine methylation of FXR1 is essential for RNA binding in cancer cells, Nucleic Acids Research , 2024;, gkae319, https://doi.org/10.1093/nar/gkae319
- Permissions Icon Permissions
Emerging evidence indicates that arginine methylation promotes the stability of arginine-glycine-rich (RGG) motif-containing RNA-binding proteins (RBPs) and regulates gene expression. Here, we report that post-translational modification of FXR1 enhances the binding with mRNAs and is involved in cancer cell growth and proliferation. Independent point mutations in arginine residues of FXR1’s nuclear export signal (R386 and R388) and RGG (R453, R455 and R459) domains prevent it from binding to RNAs that form G-quadruplex (G4) RNA structures. Disruption of G4-RNA structures by lithium chloride failed to bind with FXR1, indicating its preference for G4-RNA structure containing mRNAs. Furthermore, loss-of-function of PRMT5 inhibited FXR1 methylation both in vivo and in vitro , affecting FXR1 protein stability, inhibiting RNA-binding activity and cancer cell growth and proliferation. Finally, the enhanced crosslinking and immunoprecipitation (eCLIP) analyses reveal that FXR1 binds with the G4-enriched mRNA targets such as AHNAK, MAP1B, AHNAK2, HUWE1, DYNC1H1 and UBR4 and controls its mRNA expression in cancer cells. Our findings suggest that PRMT5-mediated FXR1 methylation is required for RNA/G4-RNA binding, which promotes gene expression in cancer cells. Thus, FXR1’s structural characteristics and affinity for RNAs preferentially G4 regions provide new insights into the molecular mechanism of FXR1 in oral cancer cells.

Dysregulated gene expression is a hallmark of cancer, and post-transcriptional gene regulation (PTR) contributes significantly to activating oncogenes and reducing tumor suppressor expression ( 1 , 2 ). The changes in PTR have gained considerable attention for their regulatory roles in biologically significant cis- and trans-factors, such as 5′- and 3′-untranslated regions (UTRs) of mRNAs and RNA-binding proteins (RBPs), respectively ( 3 ). RBPs regulate critical cellular processes, including transcription, mRNA turnover, and translation ( 4 ). However, aberrant expression of RBPs contributes to neoplasia, including head and neck oral squamous cell carcinomas ( 5 , 6 ). Although significant progress has been achieved in understanding RBP-mediated gene regulation ( 7 , 8 ), and cancer-promoting activity, the molecular basis of dysregulated expression of RBPs has yet to be studied. RBP, Fragile X mental retardation protein-1 (FXR1), is a chromosome 3q amplification gene overexpressed in multiple cancers and exerts oncogenic signaling to promote tumorigenesis ( 9–16 ). Our published findings indicate that FXR1 helps cancer cells bypass cellular senescence by stabilizing the non-coding telomerase RNA component (TERC) and destabilizing CDKN1A (p21) to promote cell growth ( 16 ). Furthermore, our findings also demonstrated that FXR1 targets p21 mRNA destabilization by recruiting miR-301a-3p in both oral and lung cancer cells ( 17 ). Although FXR1, its downstream targets, and p53/p21 pathway-mediated cellular senescence are well studied in oral and lung cancer cells, it remains unclear how elevated FXR1 protein enhances malignant transformation in cancer cells. As most RBPs undergo post-translational modifications (PTM) such as phosphorylation, acetylation, methylation, and sumoylation to regulate gene expression in cancer cells ( 18 ), here, we set out to study the impact of PTM on FXR1 and its regulatory effects on its RNA targets. Based on the observation and unproven hypothesis that FXR1 is targeted by protein methyltransferases ( 19 ), we focused on identifying and characterizing enzymes that methylate FXR1 at the post-translational level and report the functional interactions between FXR1 and methyltransferases.
For the past 30 years, several attempts have been made to understand the biological functions of Fragile-X mental retardation (FXR) proteins in Fragile-X syndrome ( 20 ). Still, a significant knowledge gap exists in appreciating the role of the FXR family of proteins in cancer cell structure, function, protein modifications, and RNA metabolism ( 21 ). The FXR family members FMRP and FXR1 contain the arginine/glycine-rich (RGG) protein domain, but FXR2 lacks the RGG domain. However, all three FXR families of proteins have K-homology domains, which are ubiquitous throughout eukaryotes ( 22 ). FXR1 contains highly conserved arginine residues in its C-terminal nuclear export signal (NES) and the RGG domain. About 0.5–1% of the total arginine residues in the human proteome are methylated and have a slow turnover rate, which will likely confer long-lasting functional properties to the target proteins ( 23 , 24 ). Adding a methyl group(s) to the arginine residues helps the proteins to interact with other proteins and nucleic acids ( 25 ). The protein arginine methyltransferases termed PRMTs (PRMT1, 3, 4 [CARM1], 5, 6 and 8), and other probable methyltransferases (PRMT2, 7, 9) are responsible for protein methylation ( 26 ). Although the RGG domain functions are relatively known, its biological significance is bypassed in the FXR family of proteins that regulate all levels of RNA metabolism ( 27–29 ). It was envisioned that the FXR1 RGG domain could be a target of arginine methyltransferases for methylation ( 30 ). However, the specific arginine methyltransferase responsible for the methylation of FXR1 has never been identified. Methylation of FMRP and FXR1 occurs mainly within their RGG box, which may influence their RNA-binding and protein-protein interactions ( 19 ). Hence, in this research, we investigated the effect of arginine methylation on FXR1’s RNA binding capacity including its specificity towards guanine rich regions in cancer cells.
FXR1 is known to be involved in mRNA transport, translational control, and mRNA binding via adenylate-uridylate-rich (AU-rich) elements (ARE) ( 31 , 32 ), and G-quartet (G4) RNA structures ( 33 , 34 ). Previous studies have shown that FXR1 undergoes distinct PTM ( 35 , 36 ). However, the enzyme responsible for FXR1’s methylation and how methylated FXR1 impacts RNA binding and alters their expression in cancer cells are unclear. For the first time, here we report, FXR1 is arginine methylated and the functional consequence of methylation relating to RNA binding activity in cancer cells. This study shows that PRMT5 interacts with FXR1 and methylates arginine at positions 386, 388, 453, 455 and 459. Interestingly, both R388 and R455 of FXR1 are necessary to bind to RNAs, with a predilection for G4-RNAs. As a result, we argue that FXR1 methylation increases its G4-RNA-binding capacity, which promotes cancer cell growth and proliferation. Furthermore, the FXR1 mRNA targets identified by nhanced crosslinking and immunoprecipitation (eCLIP)-seq had a greater binding affinity for the G4-rich sequences of top genes such AHNAK, AHNAK2, UBR4, MAP1B, DYNC1H1 and HUWE1. Studies have found that these targets have many functions in various malignancies ( 37–41 ). In addition, TCGA database analysis of HNSCC revealed amplification of these RNA targets, implying carcinogenic involvement. However, further study is required to unravel the molecular mechanism by which FXR1 regulates each of its mRNA targets to promote cancer growth. Interestingly, both genetic and small molecule PRMT5 inhibition failed to methylate recombinant as well as the endogenous FXR1, resulting in protein instability and downregulation of FXR1 target mRNA levels in HNSCC cells. Our findings explain one of the molecular mechanisms of FXR1’s reported tumorigenic role in HNSCC and lay the groundwork for future research into how targeting the interface between FXR1 and PRMT5 can affect gene expression and aid in the development of novel therapies.
Cell lines, reagents, plasmids and antibodies
HNSCC cell lines UMSCC11A, -74A and -74B were obtained from the University of Michigan, and SCC4 (#CRL-1624), SCC25 (#CRL-1628) and Cal27 (#CRL-2095) were obtained from ATCC. Lung cancer cell line A549 was also obtained from ATCC. Cell lines UMSCC74B and Cal27, and A549 were routinely grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) with 100 U/ml penicillin-streptomycin (P/S). UMSCC11A and -74A were grown in DMEM containing 10% FBS, 100 U/ml P/S, and 1X non-essential amino acids. SCC4 and SCC25 cell lines were grown in DMEM: F12 (1:1) containing 400 ng/ml hydrocortisone, 10% FBS, and 100 U/ml P/S. A549 was grown in F-12K medium containing 10% FBS and 100 U/ml P/S. shRNA constructs for FXR1 (TRCN0000158932) ( 16 , 17 ) were obtained from Sigma Mission. PRMT5 shRNA was obtained from Santa Cruz biotechnologies (SC41073-SH). Flag-PRMT5 and Flag-MEP50 were generated by cloning the corresponding cDNA into the pRK5-Flag vector ( 37 ). HA-PRMT5 was constructed by cloning the corresponding cDNA into the pRK5-HA vector ( 37 ). Myc-FXR1 was constructed by cloning the corresponding FXR1 (>NM_005087.4) into the pCDNA3.0 with C-terminal Myc-tag ( 35 ). GST-FXR1 was created by cloning (S382-P476) of FXR1 (>NM_005087.4) in the C-terminus of GST gene in pGEX-6P-1 plasmid between EcoR1 and NotI with an intervening stop codon. Single guide RNAs (sgRNA) for PRMT5 were designed at https://www.synthego.com and were cloned into lentiCRISPR v2 vector (Addgene, #52961) ( 42 , 43 ).
From Cell Signaling Technology, FXR1 (#12295, used predominantly for western blot), Myc-tag (9B11) (#2276), E-Cadherin (24E10) (#3195), N-Cadherin (D4R1H) (#13116), Symmetric Di-Methyl Arginine Motif [sdme-RG] MultiMab™ Rabbit mAb mix (#13222), Asymmetric Di-Methyl Arginine Motif [adme-R] MultiMab™ Rabbit mAb mix (#13522), CD3 (#78588S); From EMD Millipore, FXR1 (#05-1529, used for IP and RNA-IP); From Abcam, FXR1 (#ab50841 for IHC and multiplex); BD Pharmingen, p21, (#556431); From Proteintech, GAPDH (10494-1-AP), Histone H3 (17168-1-AP), GST-tag (10000-0-AP), PRMT5 (18436-1-AP), PRMT1 (11279-1-AP), HA-tag (51064-2-AP), and β-Actin (60008-1-Ig). Horseradish peroxidase-conjugated anti-mouse (NA931) and anti-rabbit (NA934) immunoglobulin Gs were procured from GE Healthcare Biosciences (Uppsala, Sweden). Normal mouse (sc-2025) and rabbit (sc-2027) IgGs were obtained from Santa Cruz Biotechnology. Protein A/G plus (sc-2003) beads were purchased from Santa Cruz Biotechnology. GSK3326593 (PRMT5) and GSK3368712 (PRMT1) inhibitors were obtained from GlaxoSmithKline (GSK) with a material transfer agreement (MTA). The protein thermal shift assay dye was procured from applied biosystems.
Senescence staining
SA-β-gal activity is determined using X-gal (5-bromo-4-chloro-3-indolyl β- d -galactoside) staining at pH 6.0 according to the manufacturer's instructions (9860, Cell Signaling Technology). A549 cells were transiently infected with Control shRNA and FXR1 shRNA for 72 h and were treated with TGFβ as described in the results section.
Immunoblot analysis
Cells were lysed using RIPA buffer, supplemented with 1× protease inhibitor cocktail and PMSF, equal amount of proteins were separated using SDS-PAGE. Proteins were transferred to the PVDF membrane, blocked in 5% skimmed milk, and incubated with primary antibodies at 4°C overnight. Membranes were washed three times with 1× Tris-buffered saline-0.1% Tween-20 and incubated with secondary antibody for 1 h at room temperature. Proteins were visualized using substrates Clarity or Clarity Max (Biorad# 1705060 and 1705062), followed by Biorad Image Lab.
Polysome profiling
A549 cells were treated with TGFβ for 48 h, cells were lysed in TMK100 lysis buffer, and the supernatant was layered onto a 10–50% sucrose gradient and centrifuged at 151 000 × g at 4°C for 3 h. Polysome fractions were collected using a fraction collector with continuous absorbance monitoring at 254 nM. RNAs were extracted with Trizol (Invitrogen) and reverse-transcribed to cDNAs using SuperScript III Reverse Transcriptase. PCR was performed using primers listed below: FXR1: 5′- CCCTAATTACACCTCCGGTTATG-3′ and 5′-TCTCCTGCCAATGACCAATC-3′; β-Actin: 5′- GGACCTGACTGACTACCTCAT-3′ and 5′-CGTAGCACAGCTTCTCCTTAAT-3′. Two percent agarose gel was utilized to resolve the PCR products. Band quantification was performed using Quantity One (Bio-Rad Laboratories, Inc.).
RNA extraction and qRT-PCR
Total RNA is prepared from HNSCC cell lines using Trizol (Ambion) or RNeasy mini kit (QIAGEN) by following the manufacturer's protocol. qRT-PCR for all m/RNA targets is performed using an Applied Biosystems StepOne Plus system or quantstudio 6.0 pro with the SYBR green master mix RT-PCR kit (SA Biosciences) as described ( 44 ). Primer sequences are provided in Supplementary Table S1 .
RNA-seq mapping and quantification
Reads were aligned to the human hg38 reference genome using STAR (v2.7.10a) ( 45 ). Genecode annotation for hg38 (version 37) was used as reference alignment annotation and downstream quantification. BAM files were filtered for uniquely mapped reads using custom bash scripts. Quality metrics were calculated using Picard tool ( http://broadinstitute.github.io/picard/ ) and summarized using MultiQC ( 46 ). Gene level expression quantification was calculated using FeatureCounts (v2.0.1) ( 47 ). Counts were calculated based on protein-coding genes from the annotation file.
Differential gene expression analysis and functional enrichment
Low-expressed genes were filtered using a per case-control approach with RPKM ≥0.5 as a filter to keep genes. Differential Expression was performed in R using DESeq2 ( 48 ). We estimated log 2 fold changes, P values, and FDR (Benjamini-Hochberg correction). We used FDR <0.05 and abs (log 2 (fold change)) ≥0.5 thresholds to define differentially expressed genes. Custom R codes were used to visualize the data. The functional annotation was performed using the R package clusterProfiler ( 49 ) using the GO database. GSEA analysis was performed using the R package fgsea. A Benjamini–Hochberg FDR was applied as a correction for multiple comparisons. Significant categories were filtered for FDR <0.05.
Transduction (shRNA or sgRNA)
Specific shRNA and control shRNA plasmids or sgRNA and controls were used for the preparation of individual lentiviral particles. Cells were transduced with the lentiviral particles at an MOI (multiplicity of infection) of 25–50 in a medium supplemented with 8 μg/ml polybrene ( 16 ) and incubated for 72 h. mRNA levels and the protein expression were analyzed by qRT-PCR and immunoblot respectively.
Purification of GST-tagged FXR1 proteins from bacteria
50 ml of log phase culture of E. coli BL21(DE3) cells containing the pGEX-6P-1-FXR1 plasmid was grown at 37°C in Luria Broth (LB) containing 100 μg/ml carbenicillin. The bacteria were induced to express human truncated FXR1 protein by adding isopropyl β- d -1-thiolgalactopyranoside (IPTG) to a final concentration of 25 uM and incubated for 4h. Cells were harvested by centrifugation at 2500 × g for 10 min at 4°C, resuspended in 10 ml lysis buffer (50 mM HEPES pH 7.9, 150 mM KCl, 1 mM MgCl2, 0.1% Triton-X 100, 0.1 mM phenylmethylsulfonylfluoride (PMSF), and Complete Protease Inhibitor Cocktail (Fisher#P178430) and lysed via sonication on ice (Fisher Scientific Sonic Dismembrator Model 100; three 10 s pulses at level 7). Debris was pelleted via centrifugation at 11 000 × g for 20 minutes at 4°C, and supernatants were added to glutathione sepharose beads for 3 h at 4°C. Beads were rocked with lysates for 1 h at 4°C, then washed 5 times with 2 ml of lysis buffer. GST-FXR1 protein was eluted by adding 0.1 ml of lysis buffer containing 50 mM reduced glutathione and a batch elution method. Eluted samples were dialyzed into a lysis buffer containing 10% glycerol and stored at −80°C.
In vitro methylation assays
PRMT5 in vitro methylation assays were performed as previously described ( 50 ). Briefly, 5 μg of recombinant GST-FXR1 proteins (WT and mutants) purified from bacterial cells were incubated with immunoprecipitated HA-PRMT5 in the presence of 1 μl of adenosyl- l -methionine, S- [methyl- 3 H] (1 mCi/ml stock solution, Perkin Elmer). The reactions were performed in the methylation buffer (50 mM Tris pH 8.5, 20 mM KCl, 10 mM MgCl 2 , 1 mM β-mercaptoethanol, and 100 mM sucrose) at 30°C for 1 h and stopped by adding 3 × SDS loading buffer and was resolved by SDS-PAGE. The separated samples were then transferred from the gel to a polyvinylidene difluoride membrane, which was then sprayed with EN 3 HANCE (Perkin Elmer) and exposed to X-ray film.
Immunoprecipitation of FXR1 from UMSCC74B cells
Endogenous FXR1 was purified from control and PRMT5 inhibitor treated 74B cells (2 × 10 6 cells). For immunoprecipitation all steps were carried out at 4°C. The cells were washed with ice-cold 1X PBS buffer followed by cell lysis using 1× cell lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na 3 VO 4 , 1 μg/ml Leupeptin and 1 mM PMSF. Lysate was incubated overnight with IP specific FXR1 or IgG anitibody followed by incubation with Dynabeads for 2 h with gentle rotation. After centrifugation, lysate was removed and beads were washed three times with 1× PBS. FXR1 was purified from the antibody-bead complex using glycine buffer (pH 2.0) and the pH of the elute was adjusted to 7.5 using Tris–HCl (pH 7.5). The protein fractions were analyzed by CBB staining and immunoblot.
Structural modelling of G4-RNA binding regions of FXR1
FXR1 region S382-P476 was modelled using Phyre2(46) and Alphafold ( 51 ) servers. As this region was seen to be completely unfolded, two peptide regions corresponding to regions 382–395 and 450–463 were separately used to thread on the FMRP peptide as seen in the PDB structure 5DE5 (in complex with G4-RNA). Mutagenesis and minimization was accomplished in Chimera ( 52 ). All models were minimized by using 1000 cycles of Steepest-descent minimization followed by 50 cycles of conjugate-gradient minimization. All atoms were kept unfixed to allow for free movement. Residue properties were kept in accordance with atom parameters defined by the AMBER ff14SB force field. Finally, hydrogen bonding interaction between G4-RNA and FXR1 peptides was mapped using the generate protocol of PDBsum1(53) hosted by EMBL-EBI. Hydrogen bonds are predicted in accordance with HBPLUS hydrogen bonding potentials developed by McDonald and Thronton ( 54 ). Figures were generated using PyMOL.
Electrophoretic mobility shift assay (EMSA)
Recombinant or endogenous FXR1 protein was assembled onto 30-mer RNA. 0.5 pmol of [y-32P] ATP or 5′ ATTO™ 550 labeled RNA was mock-treated or mixed with recombinant truncated FXR1 (WT or mutants) protein(s) and incubated at room temperature (∼25°C) for 20 min. Reactions were carried out in the final volume of 10 μl of 1X buffer containing 50 mM Tris–HCl pH 7.4, 1 mM MgCl 2 , 0.1 mM EDTA, 150 mM KCl or 150 mM LiCl 2 , 1 mM dithiothreitol (DTT) with 1 U/ul of Murine RNase Inhibitor (NEB), and 100 ug/ml BSA. After incubation, the samples were loaded onto 12% nondenaturing polyacrylamide gel containing 0.5X TBE (Tris–Cl, pH 8.0, Boric acid, EDTA). The electrophoresis was performed at room temperature in 0.5× TBE for 4 h at 125 V. The RNA distribution or shift was visualized by autoradiography after gel drying or imaging at Alexa 546 nm at fluorescence excitation.
Protein thermal shift assay
FXR1 protein stability in presence of different EMSA buffers were tested using the Protein thermal shift assay dye from applied biosystems. Each reaction was carried out in the final volume of 20 μl and the FXR1 protein melt curve was obtained in quant studio 6.0 pro using the parameters specified by the manufacturers. The raw data was analyzed to determine the normalized fluorescence value for the denatured protein using the protein thermal shift software.
eCLIP and data analysis
The eCLIP studies were performed by Eclipse Bioinnovations Inc., according to the published single-end eCLIP protocol ( 55 ). Approximately 20 million UMSCC 74B cells for two biological replicates were ultraviolet crosslinked at 400 mJ cm −2 with 254-nm radiation, cells were scrapped, washed twice with ice cold 1× PBS and stored at –80ºC until it was sent out to Eclipse Bioinnovation Inc. The RBP IP was done using eClip validated FXR1 rabbit monoclonal antibody and the library was prepared according to the published method ( 55 ). Libraries generated using the eCLIP-seq method are sequenced using standard SE50 or SE75 conditions on the Illumina HiSeq 2500 platform in standard single-end formats and peaks were compared with the size matched input (smI) and positive control. Peaks were called using the standard eCLIP processing protocol 0.2, which is available at: https://github.com/YeoLab/eclip .
Immunofluorescence
Optimized multiplex immunofluorescence was performed using the OPAL™ multiplexing method. OPAL™ is based on Tyramide Signal Amplification (TSA) using the Roche Ventana Discovery Ultra Automated Research Stainer (Roche Diagnostics, Indianapolis, IN). Tissues were stained with antibodies against [DAPI, CD3 (1:100), FXR1 (1:100), and PRMT5 (1:100)], and the fluorescence signals were generated using the following fluorophores: [OPAL dyes, Marker + Dye Pairing, Dilution used] (Akoya Biosciences, Marlborough, MA). Slides were imaged at 20X magnification using the Vectra® Polaris™ Automated Quantitative Pathology Imaging System (Akoya Biosciences, Marlborough, MA) and analyzed using inForm® Tissue Analysis Software (v[2.6.0], Akoya Biosciences, Marlborough, MA).
Cell viability and colony formation
Cell viability rate upon UMSCC74B or A549 cells treated with GSK3326593 (GSK593) and GSK3368712 (GSK712) alone or in combination for 72 h is determined using MTT cell proliferation assays (Invitrogen). Briefly, 5 × 103 cells were seeded into each well of a 96-well plate (well area = 0.32 cm 2 ). On the next day, cells were treated with 2 μM of each drug alone or in combination every 24 h, and the medium was replaced with an experimental medium (100 μl). MTT solution was prepared fresh (5 mg/ml in H2O), filtered through a 0.22-μm filter, and kept for 5 min at 37°C. The MTT solution (10 μl) was added to each well post-treatment, and plates were incubated in the dark for 4 h at 37°C. The reaction was stopped using MTT stop solution (10% SDS in 1N HCl) and further incubated overnight at 37°C. The following day the absorbance was measured at A570 nm using a plate reader (Bio-Rad).
Statistical analysis
Data are expressed as the mean ± the standard deviation. Two-sample t-tests with equal variances are used to assess differences between means. Results with P -values <0.05 are considered significant.
TGFβ-induced FXR1 undergoes post-translational modification in cancer cells
Our previous findings demonstrated that overexpressed FXR1 in metastatic oral cancer cells (UMSCC-74A, -74B) and lung adenocarcinoma A549 cells contribute to tumor growth and proliferation ( 16 , 17 ). Silencing FXR1 promotes cellular senescence by activating CDKN1A (p21) and downregulating TERC RNA in both oral and lung A549 cells ( 16 ). Hence, to determine the molecular basis of high FXR1 protein levels in cancer cells, we used A549 lung cancer cells, which show metastatic phenotype under the treatment of cytokine transforming growth factor-β (TGFβ) ( 56 ). The TGFβ-signaling mediated epithelial-to-mesenchymal transition (EMT) is a hallmark of tissue fibrosis, tumor invasiveness, and metastasis ( 57 ). Therefore, to study whether EMT plays a role in high FXR1 protein levels, we used A549 cells and tested their expression under TGFβ. As shown in Figure 1A , TGFβ induced the expression of FXR1 protein with reduced E-cadherin and increased N-cadherin levels (EMT markers). The right panel shows the FXR1 protein quantification. Although FXR1 knockdown (KD) alone showed no changes in the E-cadherin and N-cadherin levels, the addition of TGFβ in FXR1 KD cells facilitated a moderate decrease in E-cadherin and an increase in N-cadherin levels compared to only TGFβ treated cells. This observation is further confirmed by cell morphology changes, in which TGFβ-induced cells exhibit a mesenchymal phenotype and silencing of FXR1 induces senescence (Figure 1B , top panel shows quantitation of senescence). However, the changes in E- and N-cadherin levels from TGFβ treated FXR1 KD cells (Figure 1A ) may signify the changes occurring only in quiescent cells. Next, we tested whether TGFβ-induced FXR1 protein levels are mediated through transcriptional activation of the FXR1 transcript levels. Surprisingly, no difference in mRNA levels of FXR1 was observed in TGFβ-induced cells (Figure 1C ). Hence, we tested whether TGFβ influences the mRNA translation of FXR1 using a polysome gradient assay. The TGFβ-induced A549 cells showed no change in mRNA translation of FXR1 compared to untreated cells (Figure 1D ). These data indicate that high expression of FXR1 in the presence of TGFβ might be associated with post-translational modification (PTM) that may contribute to its protein stability. Therefore, we tested FXR1 protein stability by treating the cells with the protein synthesis inhibitor cycloheximide. As shown in Figure 1E and F , the TGFβ treated cells showed increased FXR1 protein stability compared to untreated cells, implying that FXR1 may undergo PTM in TGFβ-treated cells. The findings indicate that the molecular basis for overexpressed FXR1 levels in cancer cells is possibly due to PTM, which could influence its oncogenic function.

TGFβ-induced FXR1 undergoes post-translational modification in cancer cells. ( A ) Western Blot analyses show protein regulation by TGF-β treatment (48 h) on A549 cells. GAPDH serves as a loading control. The bar graph on the right side depicts the quantitative value of FXR1 in panel-A western blot. N = 3. ( B ) Analyses of cell morphology (upper panel) and β-Gal staining (lower panel) of the A549 cells treated with TGF-β and shRNA. The upper panel depicts the quantitative pixel values of β-gal positive cells, an indicator of cellular senescence. ( C ) qRT-PCR of the samples mentioned above (A and B) show that TGF-β only affects the FXR1 protein and does not affect its RNA level. N = 3. *** P < 0.0005. ( D ) Polysome profiling of A549 cells with TGF-β treatment compared to control. DNA gel shows the RT-PCR products from serial polysome fractions from control and treated TGF-β samples and analyzed for FXR1 expression in each pulled polysome fraction. ( E ) A549 cells were pretreated with TGF-β or control diluent for 48 h, followed by 5 μM cycloheximide treatment for 0 to 10 h to block protein synthesis. After the treatment, the cells were harvested at the indicated time points and immunoblotted for FXR1, P21 and β-actin (loading control). ( F ) Quantitative analyses of FXR1 protein levels in control and TGFβ treated A549 cells followed by cycloheximide treatment. The results plotted here represent the mean ± SEM of three independent experiments. All the data were defined as mean ± SD and were analyzed by Student's t -test ( n = 3). *** P < 0.0005.
PRMT5-mediated arginine methylation promotes the PTM of FXR1
The TGFβ-induced PTM of FXR1 may be carried out by phosphorylation, acetylation, or arginine methylation to promote protein stability of the RGG domain-containing proteins ( 58 ). Hence, we determined whether specific arginine methylation carried out by protein methyltransferases targets FXR1 and promotes its stability in cancer cells. We used PhosphoSitePlus (phosphosite.org) amino acid predictions and selected the methylation sites on specific arginine residues of FXR1. Based on the C-terminal NES and RGG domain amino acid sequences, we chose arginine amino acids 386, 388, 453, 455 and 459 (Figure 2A ) and studied their methylation status and interactions with different methyltransferases. The preferred amino acids are highly conserved between humans and mice and moderately conserved in a known FXR family member, FMRP (Figure 2B ), suggesting that these conserved amino acids may play a role in the biological function of FXR1 in cancer cells by contributing to its stability. To determine the arginine methyltransferase that methylates FXR1, we generated Myc-tagged FXR1 with Arg (R) to Lys (K) mutation of the above residues. We expressed and confirmed the wild-type and R mutant constructs (individually and together) in the human embryonic kidney (HEK) 293T cells (Figure 2C ). PRMT1 is the primary type I enzyme responsible for approximately 80% of total arginine methylation (asymmetric dimethylarginine [ADMA]), whereas PRMT5 is the dominant type II enzyme that generates symmetric dimethylarginine (SDMA) ( 59 ). The expression of both PRMT5 and PRMT1 has been tested in multiple head and neck squamous cell carcinoma (HNSCC) and A549 cells where OHKC (immortalized normal oral keratinocytes) and DOK (dysplastic oral keratinocytes) cells serve as normal and dysplastic cell lines ( Supplementary Figure S1A ). PRMT5 is predominantly expressed across all the cell lines compared to PRMT1. We also found that the levels of FXR1 and PRMT5 increased with TGFβ treatment ( Supplementary Figure S1B ). Hence, we tested the methylation status of wild-type (WT) and mutant FXR1. The cellular lysates from HEK 293T cells transfected with an empty vector and a plasmid expressing Myc-FXR1 (WT) were immunoprecipitated using a c-Myc antibody, separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotted for both ADMA and SDMA (Figure 2D ). An antibody specific to ADMA failed to detect methylated FXR1, however an antibody against SDMA detect the WT-FXR1 indicated that FXR1 is symmetrically dimethylated at Arg residues. HEK293T cells expressing WT and R386/459K FXR1 were subjected to immunoprecipitation (IP) with Myc-antibody and probed for anti-SDMA antibody. As shown in Figure 2E , the SDMA antibody only reacted to the WT and failed to detect any methylation on FXR1 (R386-459K), confirming the symmetrical dimethylation of these arginine residues in FXR1. Hence, to ensure PRMT5 interacts with methyl Arg residues of FXR1, both WT and R386-459K independently expressing cell lysates were subjected to IP and probed for SDMA, PRMT5, PRMT1 and a positive control FMRP (which interacts with FXR1 through N-terminal Tudor domains) ( 60 ). As shown in the figure, WT FXR1 interacts with SDMA antibody and PRMT5 through the c-terminal NES/RGG box; however, it failed to establish a strong interaction with PRMT1. This finding indicates that PRMT5 targets Arg residues of FXR1 and methylates them. More importantly, Arg residues of R388, R455 and a complete mutation of Arg residues failed to interact with PRMT5, suggesting that these two Arg residues are likely targeted by PRMT5 (Figure 2F and the bottom graph). The direct protein-protein interaction between FXR1 and PRMT5 was further confirmed using overexpressed HA-tagged PRMT5 IP lysates probed for both SDMA and FXR1 in HEK293T cells ( Supplementary Figure S1C ). Finally, we carried out the cycloheximide assay to ensure Arg residues are essential for FXR1 protein stability. Both Myc-tagged stably expressed WT and R386-459K proteins in A549 cells were treated with cycloheximide for designated times (up to 10 h) and tested for FXR1 levels by probing with Myc-Ab. As indicated in Figure 2G , after 10 hours, the WT FXR1 level is comparable to its initial time. In contrast, the mutant protein level after 10 hours was reduced to ∼50% compared to the initial time. This observation indicates that arginine residues at positions R386, 388, 453, 455 and R459 may be essential for FXR1 protein stability, individually or collectively. Thus, these observations demonstrated that PRMT5 interacts with FXR1 and promotes its stability in cancer cells.

PRMT5-mediated arginine methylation promotes PTM of FXR1. ( A ) The protein structure of FXR1 protein has regions marked for its different domains. The C-terminal arginine-glycine-glycine (RGG) RNA-binding domain has the methylated arginine (R) residues marked in the illustration. ( B ) Multiple sequence alignment of the C-terminus of human and mouse FXR1 and FMRP proteins is shown. Secondary structural elements are marked above the sequences, with α-helices depicted as cylinders and β-strands as arrows. The R residues potentially methylated inside the cell have been chosen for the mutation to lysine (K) and are highlighted (yellow). The FXR1 residue numbers are given above the sequence. The numbers in parentheses indicate the length of the sequences shown. ( C ) Immunoblot analyses of WT and mutant Myc-FXR1 protein expressions in HEK293T cells are shown. β-Actin serves as a loading control. ( D ) HEK293 cells expressing empty vector and Myc-tag FXR1 (WT) were used for IP with Myc-tag antibody and probed for SDMA, ADMA and Myc-tag antibodies. The empty vector serves as a control for Myc-FXR1. ( E ) HEK293 cells expressing empty vector, Myc-tag FXR1 (WT), and mutant (R386-459K) were used for IP with Myc-tag antibody and probed for SDMA and Myc-tag antibodies. ( F ) HEK293 cells expressing empty vector, Myc-tag FXR1 (WT), and mutants R386K, R388K, R453K, R455K and R459K were used for IP with Myc-tag antibody and probed for SDMA, PRMT5, PRMT1 and FMRP (positive control). The bottom panel depicts the quantitative value of WT and RGG mutants FXR1 protein interaction with PRMT5. N = 3. ( G ) A549 cells stably expressing Myc-tag FXR1 (WT) and mutant (R386-459K) were treated with 5 μM cycloheximide treatment for 0 to 10 h to block protein synthesis. After the treatment, the cells were harvested at the indicated time points and immunoblotted for FXR1 and β-actin (loading control). The bottom graph shows the relative FXR1 protein levels with time after cycloheximide treatment. All the data were defined as mean ± SD and were analyzed by Student's t -test ( n = 3). * P < 0.05.
Silencing PRMT5 reduces FXR1 and cell growth in HNSCC cells
PRMT5 is the primary enzyme responsible for arginine SDMA of target proteins and it prefers the consensus arginine- and glycine-rich regions known as RGG/RG motifs ( 61 ). PRMT5 targets numerous RGG domain-containing proteins, and inhibiting PRMT5 decreases target protein levels via demethylation ( 61 , 62 ). However, PRMT1 has been shown to carry out protein methylation without PRMT5, indicating a redundancy in the activation of protein methylation by these two methyltransferases ( 63 ). As a result, we investigated whether silencing PRMT5 and PRMT1 affected FXR1, FXR2 and FMRP levels in oral and lung cancer cells. As shown in Figure 3A and the right graph panel, we used two guide RNAs (CRISPR/Cas9) to knockout PRMT1 and PRMT5 in oral cancer cells (lung cancer cells, Supplementary Figure S2A ), and only PRMT5 deletion reduced FXR1 levels but not FXR2 (which lacks the RGG domain), as previously described ( 16 ). Interestingly, PRMT1-silenced cells did not change the protein levels of FXR1 or FXR2, indicating that FXR1 may be a direct substrate of PRMT5 in oral cancer cells. Furthermore, we could not detect the protein FMRP (data not shown), which is not expressed in oral or lung cancer cells.

Genetic and small-molecule inhibition of PRMT5 reduces FXR1 and cell growth in HNSCC cells. ( A ) The immunoblot shows two independent guide RNA-mediated knock out (KO) of PRMT1 and PRMT5 in UMSCC74B oral cancer cells. β-Actin serves as a loading control. Quantitative protein levels of FXR1 and FXR2 from three independent experiments are shown as a bar graph (right panel). ( B ) The panel depicts the colony-forming efficiency from clonogenicity assays of UMSCC74B cells treated with indicated drugs and DMSO for 72 h. ( C ) MTT analysis of cell viability in UMSCC74B cells treated with indicated drugs and DMSO for 72 h. Data presented as the mean ± SD of three independent experiments. ( D ) UMSCC74B cells were treated with PRMT5i and PRMT1i (1.5 μM) for 72 h. RNA extraction followed by qRT-PCR was done to determine the relative mRNA levels of FXR1, PRMT5, PRMT1 and p21. All the data were defined as mean ± SD and were analyzed by Student's t -test ( n = 3). *** P < 0.0005. ( E ) Immunoblot analysis of cell extracts obtained from UMSCC74B cells treated with PRMT5i and PRMT1i for 72 h. GAPDH serves as a loading control. The upper bar graph shows the quantitative analyses of FXR1 expression upon treatment. ( F ) Immunoblot analyses of FXR1, comparing FXR2 and PRMT5 levels in UMSCC74B cells upon PRMT5i treatment for 72 h. β-actin served as a loading control. ( G ) Endogenous FXR1 was purified from UMSCC74B control and PRMT5i (2 μM) treated cells using FXR1 specific antibody and mouse IgG (negative control) antibody. Purified protein fractions were analyzed by 10% SDS-PAGE followed by CBB staining. The bottom panel represents the immunoblot confirmation of FXR1 protein obtained from IP. ( H ) Estimating methylation status of endogenous FXR1 purified from UMSCC74B cells treated with PRMT5i (2 μM). Immunoblot was probed with FXR1 and SDMA antibody, a marker of protein methylation. ( I ) UMSCC74B cells were treated with and without PRMT5i for 72 h, followed by treatment with 5 μM cycloheximide for 0 to 8 h to block protein synthesis. After the treatment, the cells were harvested at the indicated time points and immunoblotted for FXR1 and β-actin (loading control). The bottom graph shows the relative FXR1 protein levels with time after cycloheximide treatment. N = 3. All the data were defined as mean ± SD and were analyzed by Student's t -test * P < 0.05.
Based on the effect that PRMT5 had on FXR1 levels, we wanted to see if inhibiting PRMT5 demethylated FXR1 and regulated its actions in cancer cells. GlaxoSmithKline (GSK) has found that both the PRMT1 inhibitor GSK3368712 (GSK712) and the PRMT5 inhibitor GSK3326593 (GSK593) have anti-tumor effects in a variety of cancer cell lines, with the exception of HNSCC ( 64 ). To investigate the efficacy of PRMT5 inhibition, we treated oral and lung cancer cells with single and combined PRMT5 and PRMT1 inhibitors (PRMT5/1i). The combination treatment with PRMT5/1i resulted in considerably reduced colony formation (Figure 3B and S2B) and cell growth (Figures 3C and S2C) in both cell lines. Next, we investigated the capacity of PRMT5/1i to inhibit FXR1 mRNA transcript and protein levels in cancer cells. Following the treatment described above, the mRNA and protein levels were measured in the UMSCC74B cells. In addition, we also measured the p21 levels because, FXR1 silencing was already known to regulate p21 mRNA levels ( 16 ). PRMT5i treatment had little effect on FXR1 mRNA, but it elevated p21 levels significantly in oral cancer cells (Figure 3D ). This finding suggests that FXR1 remains unchanged at the mRNA level. However, demethylation by PRMT5i could affect FXR1 protein and increase p21 levels. In addition, we checked the protein levels of FXR1 and p21 to ensure the inhibitor's effectiveness. As Figures 3E and F indicated, PRMT5 inhibition affected FXR1 but not FXR2 protein levels. Interestingly, a significant rise in p21 levels was also found in PRMT5-inhibited cells, implying that unmethylated FXR1 may be dormant in both oral and lung cancer (Figure S2D and S2E) cells. Interestingly, inhibiting PRMT1 and PRMT5 increased PARP cleavage, which can be attributed to the cell death as demonstrated by the inability to form colonies (Figure 3B ). Next, to confirm our observation that inhibiting PRMT5 methyltransferase activity reduces Arg methylation and destabilizes FXR1, we employed endogenous FXR1 isolated from control and PRMT5i-treated UMSCC74B cells (Figure 3G ). The purified fraction was tested with the SDMA antibody, which is a marker for PRMT5 activity. Our findings demonstrated that the inhibitory action of PRMT5 failed to methylate FXR1 in vivo (Figure 3H ). To investigate the effect of demethylation on FXR1 protein stability, we treated the cells with cycloheximide in PRMT5 inhibited UMSCC74B cells. The time-dependent experiment demonstrated that FXR1 protein stability is significantly reduced in PRMT5 inhibited cells, in which FXR1 is demethylated (Figure 3I and bottom panel). In addition, we wished to test whether silencing the activity of PRMT5 alters the localization of FXR1 in cancer cells. As shown in Supplementary Figure S2F , there is no change in FXR1 distribution in the cells under PRMT5 silencing condition, demonstrating that demethylation of FXR1 did not alter its cellular localization. These findings clearly showed that FXR1 is dependent on PRMT5 for its methylation and stability, and that reducing FXR1 methylation promotes p21 levels and preventing the cancer cell growth.
Arginine amino acids are essential for FXR1 to bind to G4-RNA sequences
Previous studies have shown that arginine residues in the FMRP RGG box are required for G-quadruplex (G4) RNA binding ( 19 , 65 , 66 ). As a result, we investigated whether arginine residues in FXR1 have a similar role in binding to the p21 mRNA fragment that contains a canonical G4-RNA sequence. The protein structure of FXR1 is less well-established than that of the FMRP C-terminal domain secondary structure ( 65 ). It is also unclear how FXR1 identifies G4-RNAs and which amino acids are required for binding to G4-RNAs. To assess the relevance of these arginine residues in FXR1-G4-RNA binding, we created a 30 nucleotide RNA (sequence excised from human P21 3′UTR, seg1 ( 17 )) with a G4-RNA motif (Figure 4A ). We have previously demonstrated that FXR1 binds to G4-enriched fragment of the p21 3′UTR ( 16 , 17 ). To analyze the structural workings of various arginine binding capacities, we modeled FXR1 S382-P476 using the Phyre248 and Alphafold49 servers ( 53 ). Because this region lacked any secondary structural elements, we identified two nodes for threading into G4-RNA-bound structures using the FMR1 peptide as a template (PDB ID:5DE5) ( 67 ). Here, Node1 is defined between amino acids 382- 395 (contains R386 and R388), and Node2 entails amino acids 450–463 (includes R453, R455 and R459) (Figure 4A ). Our modeling analysis showed that Node1 formed a complex with G4-RNA using R386 when threaded in either direction (from N to C terminus, Figure 4A or C to N terminus, Supplementary Figure S3A ). Specifically, R386 formed stable hydrogen bonds with G29, C30, C5, and G7 when threaded from the N to C terminus and the C to N terminus, respectively (Figure 4A ). In comparison, Node2 could only be threaded from the N to C terminus, where C to N terminus threading was disallowed due to stearic clashes of the peptide with the G4-RNA. Hence, modeling studies indicate that these two nodes are the predominant interactors of G4-RNA. Finally, we sought to determine whether FXR1 arginine amino acids are critical for binding with G4-RNA. To begin, we cloned a protein sequence comprising FXR1’s NES and RGG (S382-P476) domains in the pGEX-6P1 vector, then altered the arginine residues (R to K) and purified it using the GST-affinity purification technique ( Supplementary Figure S3B ). The in vitro methylation analysis showed that PRMT5 successfully methylated WT FXR1 but failed to adequately methylate the arginine mutants R386, R388, R45’, R459 and R386-459K (Figure 4B ), demonstrating that PRMT5 methylates arginine at these specific positions on FXR1. Further, the recombinant WT and arginine mutant FXR1 proteins were subjected to an electrophoretic mobility shift assay (EMSA) with a radiolabeled 30-mer/ fluorescently labeled G4-RNA substrate. The resulting EMSA studies showed that WT FXR1 binds with G4-RNA at a dissociation constant (Kd) of 25 nM; however, most R to K (arginine to lysine) mutants of FXR1 bind poorly with G4-RNA with high K d and the R386K, and R386/459K fails to interact with the G4-RNA (Figure 4C and D ). To validate the specificity of FXR1 to the G4 region, we used LiCl2 instead of KCl as a metal ion in the EMSA buffer and examined the binding. It has been shown that potassium stabilizes the G4-RNA over lithium ( 68 ). As shown in Figure 5A (right panel, binding curve), potassium ions enhance G4-RNA binding to FXR1. Our results showed that lithium failed to retain the G4-RNA structure and could not bind to FXR1, indicating that FXR1 prefers G4-RNA configurations. However, it is critical to demonstrate that LiCl2 does not affect FXR1 protein stability and merely destabilizes the G4 structure. As a result, we performed the protein thermal shift assay (PTSA) as described in the experimental methods. We observed that LiCl2 had no negative influence on protein stability and maintained the same melting temperature as the sample buffer containing KCl. In addition, the same trend was observed when we used the samples with RNA between different buffers (Figure 5B ). To validate our in vitro observation, we conducted the EMSA with endogenous FXR1 protein purified from UMSCC74B control and PRMT5i cells using FXR1 specific antibody. As shown in Figure 5C and the bottom panel binding curve, the endogenous FXR1 exhibited a similar binding affinity to G4-RNA in control FXR1 whereas the binding was not significant in the FXR1 purified from PRMT5i cells. Furthermore, the endogenous FXR1 lost the RNA binding when we used LiCl2 as previously seen with the recombinant protein (Figure 5D and bottom graph). Thus, our findings provide compelling evidence that FXR1 preferentially binds to G4-RNA via its selective arginine residues.
![nucleic acid research Arginine residue in the NES and RGG domain of FXR1 are essential to bind with G4-RNA sequences. (A) The sequence and plausible structure of a 30-mer RNA is used for EMSA assays. The energy-minimized model of FXR1 region 382–395 is threaded on the structure of FMR1 with G4-RNA. When threaded in either direction, R386 makes strong hydrogen bonds with G4-RNA nucleotides and backbone phosphates. Node assembly to investigate G4-RNA binding of FXR1 region 382–476. Peptides from regions 382–395 and 450–463 were used to model them with G4-RNA. Interacting arginine residues that show sensitivity to methylation are highlighted. (B) In vitro methylation assay was performed with recombinant GST-FXR1 protein purified from bacterial cells and Myc beads bound with PRMT5/MEP50. The methylation assay was carried out in the presence of 3H-SAM. The binding was performed at 4°C for 4 h, incubated with or without PIP3 (20 μM), and subjected to immunoblot analyses. PRMT5.MEP50 proteins were purified from HEK293 cells. The Ponceau stain below serves as a loading control for the immunoblot above. (C) EMSA with 5′-labeled 30-mer RNA, recombinant FXR1 (S382-P476) WT, and respective arginine mutant proteins. 0.5 pmol of [y-32P] ATP-labeled RNA was mock-treated or mixed with increasing concentrations of recombinant WT and mutant FXR1 proteins and incubated at 25°C for 20 min. Free RNA and RNP complexes are shown in the figure. (D) The binding curves and affinity constants are shown for each recombinant protein-RNA complex.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/PAP/10.1093_nar_gkae319/1/m_gkae319fig4.jpeg?Expires=1718956395&Signature=HzBvD4Fa9EZ-s1IVyq1qkc3~MtRRaFXmSkt2Zupdm5bbNe0RCGV~XAvg91zfmzOJsvSlU72Fa7GBl-31K0VmPY8gDBihwXjz1FNkc5J0bp~KAro~t8-8ZOzKaqT0cKRYbPKqC6UIS6B8WTZUEPDp4vKkOBPbbkEUCkB1O-cWpQ1EaZbmBNK4jJ6RGbogycjhMl4MzwCRdScht0Qev-T9s-mwbL5M1LZzYBZobF4UD8sAheGodCCAwRBAKk0qVfIqGWesAW9ZdB2pfgM1jDEOlk6SZVg8qTyZLUK5Lg3I-iqnHbdnG0vCZ88LLgmZjMedWJZKoiGAIVbCd6LMacn5-Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
Arginine residue in the NES and RGG domain of FXR1 are essential to bind with G4-RNA sequences. ( A ) The sequence and plausible structure of a 30-mer RNA is used for EMSA assays. The energy-minimized model of FXR1 region 382–395 is threaded on the structure of FMR1 with G4-RNA. When threaded in either direction, R386 makes strong hydrogen bonds with G4-RNA nucleotides and backbone phosphates. Node assembly to investigate G4-RNA binding of FXR1 region 382–476. Peptides from regions 382–395 and 450–463 were used to model them with G4-RNA. Interacting arginine residues that show sensitivity to methylation are highlighted. ( B ) In vitro methylation assay was performed with recombinant GST-FXR1 protein purified from bacterial cells and Myc beads bound with PRMT5/MEP50. The methylation assay was carried out in the presence of 3 H-SAM. The binding was performed at 4°C for 4 h, incubated with or without PIP3 (20 μM), and subjected to immunoblot analyses. PRMT5.MEP50 proteins were purified from HEK293 cells. The Ponceau stain below serves as a loading control for the immunoblot above. ( C ) EMSA with 5′-labeled 30-mer RNA, recombinant FXR1 (S382-P476) WT, and respective arginine mutant proteins. 0.5 pmol of [y-32P] ATP-labeled RNA was mock-treated or mixed with increasing concentrations of recombinant WT and mutant FXR1 proteins and incubated at 25°C for 20 min. Free RNA and RNP complexes are shown in the figure. ( D ) The binding curves and affinity constants are shown for each recombinant protein-RNA complex.

PRMT5-dependent FXR1 methylation is required for G4-RNA binding in HNSCC. ( A ) EMSA was performed as mentioned above with 5′ ATTO 550 labeled 30-mer RNA using recombinant WT FXR1 protein in EMSA buffer containing 150 mM KCl/LiCl 2. The RNA-protein interaction was analyzed using 10% native PAGE gel and visualized using typhoon FLA 7000 at 546 nm. The right panel shows the binding curves of EMSA. B. Protein thermal shift assay was used to screen for the effect of KCL/LiCl 2 on FXR1 using Sypro Orange. Data from protein thermal shift software show the Boltzmann (upper) and derivative (lower) melt profiles of FXR1 with or without different buffers (KCL/LiCl 2 ) , and with RNA (sample used for EMSA). Data were collected as mentioned in the methods. The median derivative T m and Boltzmann derivative T m are represented in black and green vertical lines, respectively. ( C ) EMSA was performed as indicated above with endogenous FXR1 from UMSCC74B cells with and without PRMT5 inhibitor treatment. The bottom panel represents the binding curves of EMSA. ( D ) EMSA was performed as indicated in above in a buffer containing 150 mM KCl/ LiCl 2. The bottom panel represents the binding curves of EMSA.
The RNA-binding landscape of FXR1 demonstrates its possible role in RNA regulation
In our recent findings ( 16 , 17 ), we demonstrated that FXR1 binds to the G4-specific region of p21 and degrades the mRNA in an miR301a-3p-dependent manner. In addition to our findings, others have found that FXR1 targets multiple mRNAs, including p21, in mouse C2C12 cells ( 69 ). Hence, we decided to determine the global analysis of FXR1-associated transcripts using enhanced crosslinking and immunoprecipitation (eCLIP) ( 55 ). As described, the UMSCC74B cells were subjected to UV-cross linking and IP with FXR1 for eCLIP analysis. The eCLIP followed by RNA-seq analysis (GEO: GSE252916, reviewer token- kfwrseckrlepjet), data show that FXR1 binds to diverse locations (5′ and 3′ UTR, coding and intergenic RNA regions) of several target RNAs, accounting for 21000 reproducible peaks in both biological replicates ( Supplementary Data 1 ). Further analysis revealed that 96% of FXR1 binding peaks were matched to coding sequences (Figure 6A ). However, FXR1 has also displayed a high RNA binding preference for 5′, coding, and 3′ UTR sequences (Figure 6B and the inset). Hence, both 5′ and 3′ UTR sequences were taken for further analysis due to their role in mRNA turnover and translation functions. We focused on 3′UTR sequences over 5′UTR due to their direct role in RNA turnover functions. Our data indicate that 1.86% of eCLIP peaks was also mapped on the 3′ UTR, that are highly enriched with top targets such as MAP1B, HUWE1, DYNC1H1, AHNAK2, AHNAK and UBR4. The FXR1 binding RNA sequence motifs were identified using HOMER12 de novo motif analysis ( http://homer.ucsd.edu/homer/motif/ ). Based on their P -value, the resulting motif analysis indicates that the most enriched peaks displayed high G-rich sequences (Figure 6C and Supplementary Data 2 ). Based on their G4-rich sequences and binding preference to top targets, we mapped the FXR1 binding to the respective mRNA targets using the hg19 genome browser as indicated by eCLIP analysis. As shown in Figure 6D , FXR1 IP samples showed significant enrichment of target mRNAs compared to input samples, indicating that FXR1 preferentially binds to selective regions of mRNAs. Next, we intended to determine whether the enriched mRNAs contain canonical G4-RNA sequences in their 3′UTR ( Supplementary Data 3 ). We used a G4 mapper ( 70 ) to map the potential G4 sequences in the most enriched peaks for the top FXR1 RNA targets. Surprisingly, most of the FXR1’s identified RNA targets contain numerous G4 sequences spanning from the 5′UTR to the 3′UTR ( Supplementary Data 4 ). Altogether, the findings from this eCLIP analysis further confirm our earlier in vitro and in vivo investigations, indicating FXR1 has a relatively higher affinity for binding towards G4-RNA sequences in the mRNA. Moreover, the gene ontology (GO) enrichment analysis revealed that FXR1 interacting mRNA encoding proteins are associated with cell cycle, phosphatidylinositol signaling, ubiquitin-mediated proteolysis, and nucleocytoplasmic transport ( Supplementary Data 5 ). These findings suggest that the FXR1-RNA network-associated biological processes facilitate cancer cell growth and proliferation.

RNA binding landscape of FXR1 by eCLIP and RNA seq. ( A ) The pie chart depicts the distribution of the FXR1 eCLIP peaks in the human genome analyzed from two biological replicates. UTR-untranslated region; CDS coding sequence. The data was considered with the cut-off values of peak log 2 fold enrichment ≥3 and P -value ≤0.001. ( B ) The binned FXR1 eCLIP peak coverage across all expressed genes in UMSCC74B cells. The inset represents the metagene plots of the normalized average number of peaks mapped to specific genomic regions. The 5′UTR, CDS and 3′UTR of each gene are split into 13, 100 and 70 bins, respectively. ( C ) Top ten most significantly enriched de novo sequence motifs in the FXR1-binding peaks using HOMER12. The percentage of peaks containing the discovered motifs and the p-values of the motifs calculated by a binomial test against the random genomic background was shown. ( D ) Integrated genome viewer (IGV) browser tracks the FXR1’s eCLIP peaks of top targets (based on pvalue and log 2 fold change) spanning the genomic loci of AHNAK2, MAP1B, HUWE1, UBR4, DYNC1HI and AHNAK. Detailed information about all significantly enriched eCLIP peaks can be found in Supplementary Data-1 .
Multifaceted gene regulatory roles of FXR1 in HNSCC cells
To interrogate the oncogenic functions and gene signatures essential for cancer cell growth and proliferation, we performed an RNA-seq by silencing FXR1 and PRMT5 separately using shRNAs and analyzed the high-throughput sequencing data. The silencing effect of shPRMT5 was confirmed using immunoblot ( Supplementary Figure S4A ). For this analysis, we used total RNA isolated from the WT, FXR1 KD and PRMT5 KD cells, and subjected them to bulk RNA sequencing analysis (FXR1:GSE212760, reviewer token-ypqfmuiapxetdyh, PRMT5: GSE256352, reviewer token-mdapmcyijzgjdud). Bioinformatics analyses identified several differentially expressed genes based on a threshold of q ≤ 0.05 (FDR 5%) for statistical significance and a log-fold expression change with an absolute value of at least 1. Principal Component Analyses (PCA) plot depicts the gene expression variance that is exhibited between KD samples of FXR1 and PRMT5 ( Supplementary Figure S4B ). The heat map of differentially expressed genes (DEGs) identified in the KD and control samples is depicted in Figure 7A , and S4C showed PRMT5’s DEGs. The next bar chart and the dot plot depicts the functional enrichment of DEGs from diverse biological processes in FXR1 KD (Figure 7B ) and PRMT5 KD respectively ( Supplementary Figure S4D , upregulated pathways S4E down regulated pathways). The x-axis corresponds to the number of genes in the functional ontology. The functional enrichment of FXR1 DEGs indicated top 6 hallmark gene sets obtained from the MSigDB database (Figure 7C ), demonstrating its biological importance relating to interferon pathways. More importantly, Gene Set Enrichment Analysis (GSEA) predictions, and we identified 22 pathways that FXR1 significantly impacts. The GSEA pathway further shows that several cancer pathways are negatively affected, and anti-cancer pathways are positively regulated. Graphical representation of the rank-ordered gene lists for Interferon Alfa Response and P53 Pathways hallmark gene sets (Figure 7D ). The heat-map of FXR1 KD RNA seq depicts the expression levels of various top eCLIP targets according to the highest fold change and pvalue (Figure 7E ). While analyzing the RNA-seq data of FXR1 knockdown, we observed changes in multiple pathways associated with cancer. However, examined the significance of these findings concerning the eCLIP targets of FXR1. Next, to investigate the expression of regulated mRNAs (DEGs) connected with FXR1 (eClip) under FXR1 or PRMT5 KD circumstances, we identified the mRNAs that are present in all three conditions. Specifically, 130 genes showed increased expression (Figure 7F ) and 190 genes showed decreased expression (Figure 7G ). The GO enrichment of FXR1 eCLIP target expression that is altered under FXR1 and PRMT5 KD conditions is found to be mostly enhanced in nucleic acid binding, and helicase activities and reduced in enzyme binding and regulatory activity ( Supplementary Figure S4F and S4G ). To validate the changes in FXR1-related transcripts under both KD conditions, we examined the expression of important gene targets that are tightly bound to FXR1. According to the data presented in Figure 7H , the qRT-PCR validation of selective FXR1 targets showed a predominant decrease in expression in both FXR1 and PRMT5 KD cells. Surprisingly, TCGA database analyses of HNSCC patient tissues have revealed the FXR1 top targets are altered at the mRNA level, indicating the targets may exert an oncogenic role in HNSCC ( Supplementary Figure S4H ). Moreover, the GO enrichment analyses revealed that the 18 highest-ranking mRNA targets of FXR1 are majorly involved in nitrogen metabolism, microtubule formation, axonal control, and cell proliferation (Figure 7I ). This suggests that FXR1 can bind to and stabilize these transcripts, hence possibly promoting the growth and proliferation of cancer cells. The results further indicate that the FXR1-PRMT5 axis could have a significant impact on the development of cancer through the control of the above-mentioned biological process.

FXR1 and PRMT5-dependent altered gene signatures in HNSCC cells. ( A ) Heat map of significantly differentially expressed genes identified between FXR1 KD and control samples. Rows show Z scores of normalized, log2-transformed values from differentially expressed genes (FDR < 0.05). Dendrograms depict Pearson correlation clustering of samples. ( B ) Bar plot representing the functional enrichment of FXR D1 DEGs of the top 6 genes ontology biological process (BP). The X-axis corresponds to the number of genes in the functional ontology. The Y-axis shows the top 5 functional ontologies ranked by significance. Gradient color depicts the FDR value (red = most significant, blue = least significant). ( C ) Bar plot representing the functional enrichment of FXR D1 DEGs of the top 6 hallmark gene set from MSigDB database (FDR < 0.05). The X-axis corresponds to the normalized enrichment score based on GSEA analysis. ( D ) Graphical representation of the rank-ordered gene lists for Interferon Alfa Response (NES = 3.29, FDR = 1.24e-27) and P53 Pathways (NES = 1.50, FDR = 1.27e-02) hallmark gene sets. ( E ) Heat map for the top FXR1 eCLIP RNA targets shows differential expression profile in UMSCC74B control and FXR1 KD cells. ( F ) Venn diagram represents the FXR1 eCLIP targets commonly up-regulated in both FXR1 KD and PRMT5 KD conditions. ( G ) Venn diagram represents the FXR1 eCLIP targets commonly down-regulated in both FXR1 KD and PRMT5 KD conditions. ( H ) Quantitative real-time PCR validation of top eCLIP targets having the highest fold-change and P -values compared to the size-matched input. The results plotted here represent the mean ± SEM of three independent experiments. All the data were defined as mean ± SD and were analyzed by Student's t -test ( n = 3). *** P < 0.0005. ( I ) The bar graph represents the GO enrichment analyses of the top eighteen FXR1 eCLIP targets.
Overexpressed PRMT5 and FXR1 predict poor patient outcomes and show clinical significance
Others have reported that PRMT5 is overexpressed in HNSCC ( 71 ), and inhibition of PRMT5 by EPZ015666 (GSK3235025) reduces H3K4me3-mediated Twist1 transcription and suppresses the carcinogenesis and metastasis of HNSCC ( 72 ). PRMT5 ( 73 ) and FXR1 ( 14 , 16 , 21 ) are overexpressed in multiple cancers, but combinatorial expression changes in cancers have never been reported. In addition, we tested the mRNA level changes of PRMT5 and FXR1 in The Cancer Genome Atlas (TCGA) HNSCC and lung adenocarcinoma data sets. As shown in the survival plot the overexpressed PRMT5 and FXR1 (SD > 1) alone ( Supplementary Figure S5A and S5B ) or in combination (Figure 8A ), lead to poor patient survival in HNSCC and lung cancer patients. FXR1 protein is overexpressed in oral tumors compared to normal tissue and colocalized with PRMT5, demonstrating that both proteins contribute to an oncogenic phenotype (Figure 8B ). Hence, targeting PRMT5 to modulate FXR1 functions is significant and may provide a unique anti-tumor response for HNSCC and lung adenocarcinoma patients.

PRMT5-dependent FXR1 preferentially targets oncogenes and alters its expression in HNSCC. ( A ) Kaplan–Meier plots of overall survival of stage HNSCC patients ( n = 522) stratified by FXR1 and PRMT5 mRNA expression (SD > 1). The log-rank P value and the number of cases per group are shown. ( B ) Optimized multiplex immunofluorescence showing the expression of FXR1 and PRMT5 in human HNSCC tumor and normal adjacent tissue samples. DAPI and CD3 staining was done for the nucleus and tumor markers. ( C ) Model represents the methylation dependent regulation of FXR1 and its RNA targets to promote or inhibit the tumor cell proliferation.
The results of our study have revealed that FXR1 is a target of PRMT5 for arginine methylation. Furthermore, our data indicate that arginine methylation occurs explicitly in the NES and RGG box domains of FXR1 in cancer cells. Chromosome 3q amplification in lung and oral cancer patients leads to an increase in FXR1 mRNA levels and exert oncogenic properties ( 14 , 16 ). This study has identified and added a new feature that FXR1 protein undergoes post-translational modification by PRMT5-mediated arginine methylation, which enhances the stability of FXR1 protein (Figure 1 ). Our findings also show that PRMT5 directly adds a dimethyl group to FXR1 arginine residues in cancer cells. Based on the FXR1-PRMT5 protein-protein interaction and methylation status, the residues R388K and R455K demonstrated a lack of interaction with PRMT5 compared to WT, demonstrating that these residues might have a strong preference to get methylated by PRMT5. The improved stability of FXR1 protein may be attributed to the arginine residues R388 and R455, which exhibited robust interactions with PRMT5 (Figure 2A ). Moreover, we have also demonstrated that FXR1 demethylation through inhibition of PRMT5 affected the protein stability and reduced the cancer cell proliferation (Figure 3I ).
Post-translational modifications, including arginine methylation, regulate protein functions and this modification requires approximately 12 ATPs to add a single methyl group to a protein ( 78 ). Methyl groups added to the amino groups of amino acid side chains often increase steric hindrance and reduce hydrogen bonds by replacing the amino hydrogens ( 79 ). For example, hnRNP A1 is methylated by PRMT5 on two residues, R218 and R225, which facilitates the interaction of hnRNP A1 with IRES RNA to promote IRES-dependent translation ( 82 ). Arginine methylation of different proteins, including FXR1 family protein, FMRP, affects protein–RNA interactions, protein localization, and protein-protein interactions ( 25 ). Studies have shown that the RGG box of FMRP, is known for recognizing G-quadruplex RNAs ( 81 ) and arginine residues are highly favored when it comes to RNA binding ( 80 ). Moreover, published findings showed that the folding of G4-RNAs in vitro is similar to in vivo conditions ( 83 ). For example, the sequences we used from p21 3′-UTR are folded as a G4 (Figure 4B ) to bind with FXR1 properly. Additionally, the studies have indicated that G4-RNA must be efficiently folded to interact with protein FMRP ( 84 ). Due to the close proximity of FXR1 arginine residues spanning NES and RGG motifs, there is a likelihood that PRMT5 methylates multiple arginine residues at a given time and alter the protein stability and function of FXR1. Further, this methylation also facilitates FXR1 to bind with G4-RNAs and control their expression through a potentially novel mechanism, which requires further exploration. Based on our biochemical structure prediction, we have used a 30-base RNA that forms a G4 structure to show the binding affinity of FXR1 arginine residues. Both in vitro and in vivo assays show that arginine residues present in the NES (R386 and R388) and RGG domain (R453, R455, R459) of FXR1 are essential for binding with G4-RNAs (Figures 4 and 5 ). Subsequent in vitro binding experiments using arginine mutants demonstrated that changes in arginine residues of FXR1 lead to decreased affinity for G4-RNA. Interestingly, the binding study employing the endogenous FXR1 further validated our in vitro observations and confirmed the interplay between FXR1 and PRMT5 that is vital for G4-RNA binding by FXR1 (Figure 5 ). To further prove our claim that FXR1 prefers G4-RNAs, we used LiCl2 to destabilize the G4-RNAs and see the effect through binding studies. It has been shown that structural analysis of G4-RNA with various metal ions favors potassium as a stabilizing agent over lithium ( 68 ) (Figure 4 ). Interestingly, in the presence of potassium FXR1 strongly interact with G4-RNA, but lithium destabilizes the G4-RNA structure and disrupts the binding with FXR1 (Figure 5 ), suggesting that FXR1 may prefer a noncanonical G4-structure to interact with the RNA. Previous findings from the Darnell laboratory also stated that FMRP binds with G4-RNAs and represses mRNA translation in neuronal cells ( 74 ). Thus, methylation of the arginine residues can either help increase or decrease the RNA binding capacity of the methylated protein.
Our published findings show that FXR1 specifically targets the G4-rich regions of p21 mRNA and TERC long non-coding RNA to control their expression in oral cancer cells ( 16 ). Deleting the G4-region of p21 mRNA specifically did not interact with FXR1 in cancer cells, indicating that FXR1 prefers G4-sequences in the 3′UTR to regulate the expression of target genes. FXR1 facilitates the degradation of p21 mRNA at the molecular level by enlisting miR-133a-3p and PNPase to induce instability ( 17 ). The mechanism by which FXR1 binds to and stabilizes TERC RNA through interaction with the G4-region is not well understood. TERC RNA may not have miRNA binding sites, hence FXR1 interaction could potentially enhance TERC stability rather than destabilize it. Darnell group showed that FMRP interacts with the coding region of many mRNAs associated with autism spectrum disorders ( 75 ). Interestingly, FMRP is known to interact with G4-RNA sequences located at the 3′UTR, influencing the localization and translation of target mRNAs ( 77 ). It is also vital to show in this study that FXR1 prefers the G4-mRNAs in head and neck cancer cells, mostly localized in the cytoplasm. Nevertheless, our eCLIP data clearly demonstrate that FXR1 interacts with and regulates the target mRNAs both in a positive and negative manner in cancer cells (Figure 6 ). Utilizing the eCLIP analysis and FXR1 KD gene signature analysis, we have successfully demonstrated that the differential gene expression is mediated by FXR1. According to the eCLIP motif analysis, FXR1 can bind to both G- and U-rich sequences. The FXR1 target mRNA encoding proteins include AHNAK, AHNAK2, MAP1B, HUWE1, and DYNC1H1, as depicted in Figure 6 , enriched with G4-sequences. Our data also show that FXR1 targets the coding regions, 5′UTR, and 3′UTR of key genes involved in microtubule filaments, potentially linked to cancer progression (Figure 7 ). For instance, MAP1B, a microtubule filament protein, the prominent target of FXR1, is also targeted by FMRP and is associated with autistic spectrum disorder and autophagy ( 76 ). Therefore, establishing the connection between FXR1 and the microtubule-associated gene network would reveal the crucial role of FXR1 in cancer cells. Further experimental strategies are needed to determine if FXR1 binds to non-G4 RNAs and acts as a repressor or promoter of their mRNA turnover and translation in cancer cells.
Our RNA-seq and eCLIP analysis showed that silencing FXR1 can have both cancer positive and negative effects on gene expression, suggesting that the recognition of G4-region may influence mRNA turnover regulation. The contrasting roles of FXR1 in mRNA stability and destabilization considering the G4-structural features need to be investigated further in cancer cells. Together, our results show that arginine methylation may influence its target mRNAs having preference towards G4 enriched sequences to regulate its gene expression in cancer cells. FXR1 shows high methylation levels and can have more preference to bind G4-RNAs containing regulatory signals for generating proteins that are crucial for encouraging tumor growth. Thus, the current results indicate a straightforward function of FXR1 in cancer cells that may pave the way for targeting the NES/RGG box for therapeutic intervention to elucidate the regulation of tumor suppressors in cancer cells.
Collectively, our data unambiguously demonstrated the molecular interaction between PRMT5 and FXR1 by the impartial techniques. As demonstrated in Figure 8 , head and neck tumors have limited survival and poor outcomes due to the overexpression of FXR1 and PRMT5. The rationale behind integrating FXR1 and PRMT5 inhibitors to improve clinical outcomes is presented in our work. More importantly, as our model illustrates (Figure 8C ), we showed that PRMT5-activated FXR1 is intricate in controlling the mRNA expression of its targets, playing both tumor-activating and tumor-suppressive roles. Therefore, further research is required to fully comprehend FXR1’s involvement in mRNA synthesis and turnover in cancer cells, leading to cancer growth and proliferation.
The data underlying this article are available in the Gene Expression Omnibus, and can be accessed under accession codes GSE252916, GSE212760 and GSE256352. Further data is available in ModelArchive at https://modelarchive.org/doi/10.5452/ma-epklf .
Supplementary Data are available at NAR Online.
This work was supported by the National Institutes of Health NIH Grant R01 DE030013 and R21DE032461. Supported in part by the Translational Science Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313). This study received funding from the UNM Comprehensive Cancer Center assistance Grant NCI P30CA118100. The study also utilized the Analytical and Translational Genomics Shared Resource at University of New Mexico, which receives financial assistance from the State of New Mexico.
National Institutes of Health [R01DE030013, R21DE032461]. Funding for open access charge: NIH.
Conflict of interest statement . None declared.
Choi P.S. , Thomas-Tikhonenko A. RNA-binding proteins of COSMIC importance in cancer . J. Clin. Invest. 2021 ; 131 : e151627 .
Google Scholar
Qin H. , Ni H. , Liu Y. , Yuan Y. , Xi T. , Li X. , Zheng L. RNA-binding proteins in tumor progression . J. Hematol. Oncol. 2020 ; 13 : 90 .
Gehring N.H. , Wahle E. , Fischer U. Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation . Trends Biochem. Sci. 2017 ; 42 : 369 – 382 .
Glisovic T. , Bachorik J.L. , Yong J. , Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation . FEBS Lett. 2008 ; 582 : 1977 – 1986 .
Pereira B. , Billaud M. , Almeida R. RNA-binding proteins in cancer: old players and new actors . Trends Cancer . 2017 ; 3 : 506 – 528 .
Palanisamy V. , Jakymiw A. , Van Tubergen E.A. , D'Silva N.J. , Kirkwood K.L Control of cytokine mRNA expression by RNA-binding proteins and microRNAs . J. Dent. Res. 2012 ; 91 : 651 – 658 .
Hentze M.W. , Castello A. , Schwarzl T. , Preiss T. A brave new world of RNA-binding proteins . Nat. Rev. Mol. Cell Biol. 2018 ; 19 : 327 – 341 .
Corley M. , Burns M.C. , Yeo G.W. How RNA-binding proteins interact with RNA: molecules and mechanisms . Mol. Cell . 2020 ; 78 : 9 – 29 .
Cao H. , Gao R. , Yu C. , Chen L. , Feng Y. The RNA-binding protein FXR1 modulates prostate cancer progression by regulating FBXO4 . Funct. Integr. Genomics . 2019 ; 19 : 487 – 496 .
Cao S. , Zheng J. , Liu X. , Liu Y. , Ruan X. , Ma J. , Liu L. , Wang D. , Yang C. , Cai H. et al. . FXR1 promotes the malignant biological behavior of glioma cells via stabilizing MIR17HG . J. Exp. Clin. Cancer Res. 2019 ; 38 : 37 .
Jo Y.S. , Kim S.S. , Kim M.S. , Yoo N.J. , Lee S.H. Frameshift mutation of FXR1 encoding a RNA-binding protein in gastric and colorectal cancers with microsatellite instability . Pathol. Oncol. Res. 2017 ; 23 : 453 – 454 .
Mao L. , Tang Y. , Deng M.J. , Huang C.T. , Lan D. , Nong W.Z. , Li L. , Wang Q. A combined biomarker panel shows improved sensitivity and specificity for detection of ovarian cancer . J. Clin. Lab. Anal. 2022 ; 36 : e24232 .
Phelps H.M. , Pierce J.M. , Murphy A.J. , Correa H. , Qian J. , Massion P.P. , Lovvorn H.N. 3rd FXR1 expression domain in Wilms tumor . J. Pediatr. Surg. 2019 ; 54 : 1198 – 1205 .
Qian J. , Hassanein M. , Hoeksema M.D. , Harris B.K. , Zou Y. , Chen H. , Lu P. , Eisenberg R. , Wang J. , Espinosa A. et al. . The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers . Proc. Natl. Acad. Sci. U.S.A. 2015 ; 112 : 3469 – 3474 .
Xu M.C. , Ghani M.O. , Apple A. , Chen H. , Whiteside M. , Borinstein S.C. , Correa H. , Lovvorn H.N. Changes in FXR1 expression after chemotherapy for Rhabdomyosarcoma . J. Pediatr. Surg. 2021 ; 56 : 1148 – 1156 .
Majumder M. , House R. , Palanisamy N. , Qie S. , Day T.A. , Neskey D. , Diehl J.A. , Palanisamy V. RNA-binding protein FXR1 regulates p21 and TERC RNA to bypass p53-mediated cellular senescence in OSCC . PLoS Genet. 2016 ; 12 : e1006306 .
Majumder M. , Palanisamy V. RNA binding protein FXR1-miR301a-3p axis contributes to p21WAF1 degradation in oral cancer . PLoS Genet. 2020 ; 16 : e1008580 .
Velazquez-Cruz A. , Banos-Jaime B. , Diaz-Quintana A. , De la Rosa M.A. , Diaz-Moreno I. Post-translational control of RNA-binding proteins and disease-related dysregulation . Front Mol. Biosci. 2021 ; 8 : 658852 .
Stetler A. , Winograd C. , Sayegh J. , Cheever A. , Patton E. , Zhang X. , Clarke S. , Ceman S. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp . Hum. Mol. Genet. 2006 ; 15 : 87 – 96 .
Santoro M.R. , Bray S.M. , Warren S.T. Molecular mechanisms of fragile X syndrome: a twenty-year perspective . Annu. Rev. Pathol. 2012 ; 7 : 219 – 245 .
Majumder M. , Johnson R.H. , Palanisamy V. Fragile X-related protein family: a double-edged sword in neurodevelopmental disorders and cancer . Crit. Rev. Biochem. Mol. Biol. 2020 ; 55 : 409 – 424 .
Ozdilek B.A. , Thompson V.F. , Ahmed N.S. , White C.I. , Batey R.T. , Schwartz J.C. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding . Nucleic Acids Res. 2017 ; 45 : 7984 – 7996 .
Bulau P. , Zakrzewicz D. , Kitowska K. , Wardega B. , Kreuder J. , Eickelberg O. Quantitative assessment of arginine methylation in free versus protein-incorporated amino acids in vitro and in vivo using protein hydrolysis and high-performance liquid chromatography . BioTechniques . 2006 ; 40 : 305 – 310 .
Uhlmann T. , Geoghegan V.L. , Thomas B. , Ridlova G. , Trudgian D.C. , Acuto O. A method for large-scale identification of protein arginine methylation . Mol. Cell. Proteomics . 2012 ; 11 : 1489 – 1499 .
Bedford M.T. , Clarke S.G. Protein arginine methylation in mammals: who, what, and why . Mol. Cell . 2009 ; 33 : 1 – 13 .
Bedford M.T. Arginine methylation at a glance . J. Cell Sci. 2007 ; 120 : 4243 – 4246 .
Jarvelin A.I. , Noerenberg M. , Davis I. , Castello A. The new (dis)order in RNA regulation . Cell Commun. Signal. 2016 ; 14 : 9 .
Gerstberger S. , Hafner M. , Tuschl T. A census of human RNA-binding proteins . Nat. Rev. Genet. 2014 ; 15 : 829 – 845 .
Rajyaguru P. , Parker R. RGG motif proteins: modulators of mRNA functional states . Cell Cycle . 2012 ; 11 : 2594 – 2599 .
Ong S.E. , Mittler G. , Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC . Nat. Methods . 2004 ; 1 : 119 – 126 .
Garnon J. , Lachance C. , Di Marco S. , Hel Z. , Marion D. , Ruiz M.C. , Newkirk M.M. , Khandjian E.W. , Radzioch D Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level . J. Biol. Chem. 2005 ; 280 : 5750 – 5763 .
Vasudevan S. , Steitz J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2 . Cell . 2007 ; 128 : 1105 – 1118 .
Schaeffer C. , Bardoni B. , Mandel J.L. , Ehresmann B. , Ehresmann C. , Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif . EMBO J. 2001 ; 20 : 4803 – 4813 .
Bechara E. , Davidovic L. , Melko M. , Bensaid M. , Tremblay S. , Grosgeorge J. , Khandjian E.W. , Lalli E. , Bardoni B. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure . Nucleic Acids Res. 2007 ; 35 : 299 – 306 .
Qie S. , Majumder M. , Mackiewicz K. , Howley B.V. , Peterson Y.K. , Howe P.H. , Palanisamy V. , Diehl J.A. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma . Nat. Commun. 2017 ; 8 : 1534 .
Say E. , Tay H.G. , Zhao Z.S. , Baskaran Y. , Li R. , Lim L. , Manser E. A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1 . Mol. Cell . 2010 ; 38 : 236 – 249 .
Inoue H. , Kanda T. , Hayashi G. , Munenaga R. , Yoshida M. , Hasegawa K. , Miyagawa T. , Kurumada Y. , Hasegawa J. , Wada T. et al. . A MAP1B-cortactin-Tks5 axis regulates TNBC invasion and tumorigenesis . J. Cell Biol. 2024 ; 223 : e202303102 .
Lee C. , Park S.H. , Yoon S.K. The E3 ligase HUWE1 increases the sensitivity of CRC to oxaliplatin through TOMM20 degradation . Oncogene. 2024 ; 43 : 636 – 649 .
Leli N.M. , Koumenis C. A novel ubiquitin complex regulates aneuploid epithelial tumors by moderating an integrated stress response . Cancer Discov. 2023 ; 13 : 535 – 537 .
Wang Y. , Han J. , Zhou H. , Ai S. , Wan D A prognosis marker dynein cytoplasmic 1 heavy chain 1 correlates with EMT and immune signature in liver hepatocellular carcinoma by bioinformatics and experimental analysis . Dis. Markers . 2022 ; 2022 : 6304859 .
Zhang S. , Cai Z. , Li H. AHNAKs roles in physiology and malignant tumors . Front. Oncol. 2023 ; 13 : 1258951 .
Sanjana N.E. , Shalem O. , Zhang F. Improved vectors and genome-wide libraries for CRISPR screening . Nat. Methods . 2014 ; 11 : 783 – 784 .
Yin S. , Liu L. , Brobbey C. , Palanisamy V. , Ball L.E. , Olsen S.K. , Ostrowski M.C. , Gan W. PRMT5-mediated arginine methylation activates AKT kinase to govern tumorigenesis . Nat. Commun. 2021 ; 12 : 3955 .
Talwar S. , Jin J. , Carroll B. , Liu A. , Gillespie M.B. , Palanisamy V. Caspase-mediated cleavage of RNA-binding protein HuR regulates c-Myc protein expression after hypoxic stress . J. Biol. Chem. 2011 ; 286 : 32333 – 32343 .
Dobin A. , Davis C.A. , Schlesinger F. , Drenkow J. , Zaleski C. , Jha S. , Batut P. , Chaisson M. , Gingeras T.R. STAR: ultrafast universal RNA-seq aligner . Bioinformatics . 2013 ; 29 : 15 – 21 .
Ewels P. , Magnusson M. , Lundin S. , Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report . Bioinformatics . 2016 ; 32 : 3047 – 3048 .
Liao Y. , Smyth G.K. , Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features . Bioinformatics . 2014 ; 30 : 923 – 930 .
Anders S. , Huber W. Differential expression analysis for sequence count data . Genome Biol. 2010 ; 11 : R106 .
Yu G. , Wang L.G. , Han Y. , He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters . OMICS . 2012 ; 16 : 284 – 287 .
Cheng D. , Vemulapalli V. , Bedford M.T. Methods applied to the study of protein arginine methylation . Methods Enzymol. 2012 ; 512 : 71 – 92 .
Jumper J. , Evans R. , Pritzel A. , Green T. , Figurnov M. , Ronneberger O. , Tunyasuvunakool K. , Bates R. , Žídek A. , Potapenko A. et al. . Highly accurate protein structure prediction with AlphaFold . Nature . 2021 ; 596 : 583 – 589 .
Pettersen E.F. , Goddard T.D. , Huang C.C. , Couch G.S. , Greenblatt D.M. , Meng E.C. , Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis . J. Comput. Chem. 2004 ; 25 : 1605 – 1612 .
Laskowski R.A. , Jabłońska J. , Pravda L. , Vařeková R.S. , Thornton J.M. PDBsum: structural summaries of PDB entries . Protein Sci. 2018 ; 27 : 129 – 134 .
McDonald I.K. , Thornton J.M. Satisfying hydrogen bonding potential in proteins . J. Mol. Biol. 1994 ; 238 : 777 – 793 .
Van Nostrand E.L. , Pratt G.A. , Shishkin A.A. , Gelboin-Burkhart C. , Fang M.Y. , Sundararaman B. , Blue S.M. , Nguyen T.B. , Surka C. , Elkins K. et al. . Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) . Nat. Methods . 2016 ; 13 : 508 – 514 .
Kawata M. , Koinuma D. , Ogami T. , Umezawa K. , Iwata C. , Watabe T. , Miyazono K. TGF-beta-induced epithelial-mesenchymal transition of A549 lung adenocarcinoma cells is enhanced by pro-inflammatory cytokines derived from RAW 264.7 macrophage cells . J. Biochem. 2012 ; 151 : 205 – 216 .
Kalluri R. , Weinberg R.A. The basics of epithelial-mesenchymal transition . J. Clin. Invest. 2009 ; 119 : 1420 – 1428 .
Guccione E. , Richard S. The regulation, functions and clinical relevance of arginine methylation . Nat. Rev. Mol. Cell Biol. 2019 ; 20 : 642 – 657 .
Blanc R.S. , Richard S. Arginine methylation: the coming of age . Mol. Cell . 2017 ; 65 : 8 – 24 .
Siomi M.C. , Zhang Y. , Siomi H. , Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them . Mol. Cell. Biol. 1996 ; 16 : 3825 – 3832 .
Thandapani P. , O’Connor T.R. , Bailey T.L. , Richard S. Defining the RGG/RG motif . Mol. Cell . 2013 ; 50 : 613 – 623 .
Mersaoui S.Y. , Yu Z. , Coulombe Y. , Karam M. , Busatto F.F. , Masson J.Y. , Richard S. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids . EMBO J. 2019 ; 38 : e100986 .
Gao G. , Zhang L. , Villarreal O.D. , He W. , Su D. , Bedford E. , Moh P. , Shen J. , Shi X. , Bedford M.T. et al. . PRMT1 loss sensitizes cells to PRMT5 inhibition . Nucleic Acids Res. 2019 ; 47 : 5038 – 5048 .
Fedoriw A. , Rajapurkar S.R. , O’Brien S. , Gerhart S.V. , Mitchell L.H. , Adams N.D. , Rioux N. , Lingaraj T. , Ribich S.A. , Pappalardi M.B. et al. . Anti-tumor activity of the Type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss . Cancer Cell . 2019 ; 36 : 100 – 114 .
Phan A.T. , Kuryavyi V. , Darnell J.C. , Serganov A. , Majumdar A. , Ilin S. , Raslin T. , Polonskaia A. , Chen C. , Clain D. et al. . Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction . Nat. Struct. Mol. Biol. 2011 ; 18 : 796 – 804 .
Vasilyev N. , Polonskaia A. , Darnell J.C. , Darnell R.B. , Patel D.J. , Serganov A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP . Proc. Natl. Acad. Sci. U.S.A. 2015 ; 112 : E5391 – E5400 .
Vasilyev N. , Polonskaia A. , Darnell J.C. , Darnell R.B. , Patel D.J. , Serganov A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP . Proc. Natl. Acad. Sci. U.S.A. 2015 ; 112 : E5391 – E5400 .
Kwok C.K. , Sahakyan A.B. , Balasubramanian S. Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA . Angew. Chem. Int. Ed. Engl. 2016 ; 55 : 8958 – 8961 .
Davidovic L. , Durand N. , Khalfallah O. , Tabet R. , Barbry P. , Mari B. , Sacconi S. , Moine H. , Bardoni B. A novel role for the RNA-binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability . PLoS Genet. 2013 ; 9 : e1003367 .
Kikin O. , D’Antonio L. , Bagga P.S. QGRS mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences . Nucleic Acids Res. 2006 ; 34 : W676 – W682 .
Amano Y. , Matsubara D. , Yoshimoto T. , Tamura T. , Nishino H. , Mori Y. , Niki T. Expression of protein arginine methyltransferase-5 in oral squamous cell carcinoma and its significance in epithelial-to-mesenchymal transition . Pathol. Int. 2018 ; 68 : 359 – 366 .
Fan Z. , He L. , Li M. , Cao R. , Deng M. , Ping F. , Liang X. , He Y. , Wu T. , Tao X. et al. . Targeting methyltransferase PRMT5 retards the carcinogenesis and metastasis of HNSCC via epigenetically inhibiting Twist1 transcription . Neoplasia . 2020 ; 22 : 617 – 629 .
Xiao W. , Chen X. , Liu L. , Shu Y. , Zhang M. , Zhong Y. Role of protein arginine methyltransferase 5 in human cancers . Biomed. Pharmacother. 2019 ; 114 : 108790 .
Darnell J.C. , Jensen K.B. , Jin P. , Brown V. , Warren S.T. , Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function . Cell . 2001 ; 107 : 489 – 499 .
Darnell J.C. , Van Driesche S.J. , Zhang C. , Hung K.Y. , Mele A. , Fraser C.E. , Stone E.F. , Chen C. , Fak J.J. , Chi S.W. et al. . FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism . Cell . 2011 ; 146 : 247 – 261 .
Guo Y. , Shen M. , Dong Q. , Mendez-Albelo N.M. , Huang S.X. , Sirois C.L. , Le J. , Li M. , Jarzembowski E.D. , Schoeller K.A. et al. . Elevated levels of FMRP-target MAP1B impair human and mouse neuronal development and mouse social behaviors via autophagy pathway . Nat. Commun. 2023 ; 14 : 3801 .
Goering R. , Hudish L.I. , Guzman B.B. , Raj N. , Bassell G.J. , Russ H.A. , Dominguez D. , Taliaferro J.M. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences . eLife . 2020 ; 9 : e52621 .
Gary J.D. , Clarke S. RNA and protein interactions modulated by protein arginine methylation . Prog. Nucleic Acid Res. Mol. Biol. 1998 ; 61 : 65 – 131 .
McBride A.E. , Silver P.A. State of the arg: protein methylation at arginine comes of age . Cell . 2001 ; 106 : 5 – 8 .
Jones S. , Daley D.T. , Luscombe N.M. , Berman H.M. , Thornton J.M. Protein-RNA interactions: A structural analysis . Nucleic Acids Res. 2001 ; 29 : 943 – 954 .
Darnell J.C. , Fraser C.E. , Mostovetsky O. , Darnell R.B. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs . Hum. Mol. Genet. 2009 ; 18 : 3164 – 3177 .
Gao G. , Dhar S. , Bedford M.T. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1 . Nucleic Acids Res. 2017 ; 45 : 4359 – 4369 .
Guo J.U. , Bartel D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria . Science . 2016 ; 353 : aaf5371 .
Subramanian M. , Rage F. , Tabet R. , Flatter E. , Mandel J.L. , Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting . EMBO Rep. 2011 ; 12 : 697 – 704 .
Author notes
Email alerts, citing articles via.
- Editorial Board
Affiliations
- Online ISSN 1362-4962
- Print ISSN 0305-1048
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

IMAGES
VIDEO
COMMENTS
The 2023 Nucleic Acids Research Web Server issue is the 21st in a series of annual issues dedicated to web-based software resources for analysis and visualization of molecular biology data. This issue includes 81 articles covering web servers that support research activities in a wide range of areas, ranging from software aimed at the wet lab ...
Browse the latest articles on nucleic acid research, covering topics such as gene regulation, chromatin, epigenetics, miRNA, DNA repair and more. Find abstracts, lay summaries, supplementary data and full text links for each article.
Abstract. Nucleic acids, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), carry genetic information which is read in cells to make the RNA and proteins by which living things function. The well-known structure of the DNA double helix allows this information to be copied and passed on to the next generation.
Other research demonstrated that viruses could specifically infect and thus regress tumours 135,136 and that injecting certain tissues with nucleic acids could produce genes of interest 137 ...
Nucleic acids function in encoding, transmitting and expressing genetic information in either the double-stranded form (mostly for DNA) or in single-stranded form (mostly for RNA). Latest Research ...
Read the latest Research articles in Nucleic acids from Nature. ... Four different XNAs — polymers with backbone chemistries not found in nature, namely, arabino nucleic acids, 2 ...
Nucleic Acids Research (NAR) publishes the results of leading edge research into physical, chemical, biochemical and biological aspects of nucleic acids and proteins involved in nucleic acid ...
NEW AND UPDATED DATABASES. The 28th annual Nucleic Acids Research Database Issue contains 189 papers spanning, as usual, a wide range of biology. Unsurprisingly, COVID-19 casts a long shadow over the Issue. Seven new databases specifically address the pandemic and the SARS-CoV-2 virus responsible (Table (Table1) 1) but new and returning databases in all areas have rushed to support research ...
Examining chromatin heterogeneity through PacBio long-read sequencing of M.EcoGII methylated genomes: an m 6 A detection efficiency and calling bias correcting pipeline
In conclusion, this review provided insights into future trends and challenges in nucleic acid research, highlighting the enormous potential and impact of this field. Developments in these areas will not only deepen our understanding of fundamental biological processes but also pave the way for new diagnostic tools, personalized medicine, and ...
Nucleic Acids Research is an open-access peer-reviewed scientific journal published since 1974 by the Oxford University Press. The journal covers research on nucleic acids, such as DNA and RNA, and related work. According to the Journal Citation Reports, the journal's 2021 impact factor is 19.160.
This review explores the applications of nucleic acids and oligonucleotides in biosensing, gene regulation, drug delivery, and therapy. It covers the properties, modifications, and challenges of these biomaterials and tools in biomedical research and innovation.
Learn about nucleic acid, the main information-carrying molecule of the cell, and its two main classes: DNA and RNA. Explore the chemistry, biosynthesis, and functions of nucleic acids, as well as their role in protein synthesis and heredity.
NEW AND UPDATED DATABASES. In its 30th incarnation, the Nucleic Acids Research Database Issue once again ranges across biology with a total of 178 papers. Table Table1 1 lists the 90 new databases included, a recent record number, and there are 82 update papers from resources previously covered by NAR. Finally, six databases most recently published elsewhere contribute updates (Table (Table2). 2).
At Environmental Science & Technology (ES&T) and Environmental Science & Technology Letters (ES&T Letters), we receive large numbers of papers utilizing nucleic-acid sequencing tools, but these papers do not always effectively address research questions or evaluate hypotheses. Most such manuscripts are rejected without sending them out for peer ...
Nucleic acid sensing is involved in viral infections, immune response-related diseases, and therapeutics. Based on the composition of nucleic acids, nucleic acid sensors are defined as DNA or RNA ...
The 2021 Nucleic Acids Research database Issue contains 189 papers spanning a wide range of biological fields and investigation. It includes 89 papers reporting on new databases and 90 covering recent changes to resources previously published in the Issue. A further ten are updates on databases most …
Genome-Informed Awareness System to Examine Cancer-Derived Raw HTS Outputs Using Electronic-Probe Diagnostic Nucleic-acid Analysis (EDNA) About 606,880 Americans were expected to die of cancer in 2019, which translates to about 1,660 deaths per day, and more than 1.7 million new cancer cases were expected to be diagnosed in 2019.
The 29th annual Nucleic Acids Research Database Issue contains 185 papers covering topics from across biology and beyond. The ongoing COVID-19 pandemic continues to play a major role, inspiring the construction of seven new databases (Table (Table1). 1). The reader will also find its impact obvious in papers describing other new and returning ...
The goal is to control cationic copolymer binding affinity with anionic charged nucleic acids by controlling the monomer functionality and distribution along the polymer backbone. The resulting polyplexes will be characterized via static light scattering, dynamic light scattering, transmission electron microscopy and their stability in salt and ...
Here, Bou-Nader et al., define the nucleic acid-binding specificity of S9.6 and report its crystal structures free and bound to a hybrid, which reveal the asymmetric recognition of the RNA and DNA ...
The "Nucleic Acid Sample Preparation Market - A Global and Regional Analysis: Focus on Technology, Application, Product, End User, and Country - Analysis and Forecast, 2023-2033" report has been added to ResearchAndMarkets.com's offering.. The global nucleic acid sample preparation market is projected to reach $5,615.9 million by 2033 from $2,922.8 million in 2023, growing at a CAGR of 6.75% ...
Nucleic Acids Res; Nucleic Acids Research Vols. 1 to 52; 1974 to 2024; Vol. 52 2024: v.52(D1): D1-D1702 2024 Jan 5: v.52(1): 1-509 2024 Jan 11: v.52(2): 511-1003 ... Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press. Follow NCBI. Connect with NLM National Library of Medicine 8600 Rockville Pike ...
The GO enrichment of FXR1 eCLIP target expression that is altered under FXR1 and PRMT5 KD conditions is found to be mostly enhanced in nucleic acid binding, and helicase activities and reduced in enzyme binding and regulatory activity (Supplementary Figure S4F and S4G). To validate the changes in FXR1-related transcripts under both KD ...