- Search by keyword
- Search by citation
Page 1 of 24

Association of interleukin-17A and chemokine/vascular endothelial growth factor-induced angiogenesis in newly diagnosed patients with bladder cancer
The human interleukin-17 (IL-17) family comprises IL-17A to IL-17 F; their receptors are IL-17RA to IL-17RE. Evidence revealed that these cytokines can have a tumor-supportive or anti-tumor impact on human mal...
- View Full Text
Causal relationship between immune cells and telomere length: mendelian randomization analysis
The causal relationship between immune cells and telomere length remains controversial.
Subcutaneous immunoglobulin replacement therapy in patients with immunodeficiencies – impact of drug packaging and administration method on patient reported outcomes
Here, the perspective of patients with primary and secondary immunodeficiency receiving subcutaneous immunoglobulin (SCIg) via introductory smaller size pre-filled syringes (PFS) or vials were compared.
Dendritic cells under allergic condition enhance the activation of pruritogen-responsive neurons via inducing itch receptors in a co-culture study
Itch sensitization has been reported in patients with chronic allergic skin diseases and observed in a mouse model of allergic contact dermatitis (ACD). There is evidence suggesting that neuroimmune interactio...
Network pharmacology-based strategy to investigate the mechanisms of artemisinin in treating primary Sjögren’s syndrome
The study aimed to explore the mechanism of artemisinin in treating primary Sjögren’s syndrome (pSS) based on network pharmacology and experimental validation.
Hyperactivation and enhanced cytotoxicity of reduced CD8 + gamma delta T cells in the intestine of patients with Crohn’s disease correlates with disease activity
We aimed to investigate the immune characteristics of intestinal CD8 + gamma delta T (CD8 + γδ T) cells in Crohn’s disease (CD) and their correlation with disease activity.
Immune responses to P falciparum antibodies in symptomatic malaria patients with variant hemoglobin genotypes in Ghana
Haemoglobin (Hb) variants such as sickle cell trait (SCT/HbAS) play a role in protecting against clinical malaria, but little is known about the development of immune responses against malaria parasite ( Plasmodiu...
Systematic evaluation of B-cell clonal family inference approaches
The reconstruction of clonal families (CFs) in B-cell receptor (BCR) repertoire analysis is a crucial step to understand the adaptive immune system and how it responds to antigens. The BCR repertoire of an ind...
Expression of the immune checkpoint molecules CD226 and TIGIT in preeclampsia patients
Imbalanced immune responses are involved in developing preeclampsia (PE). We wish to explore the expression and potential changes of immune checkpoint molecules TIGIT, CD226 and CD155 in PE patients.
Oral administration of DNA alginate nanovaccine induced immune-protection against Helicobacter pylori in Balb/C mice
Helicobacter pylori (H. Pylori), is an established causative factor for the development of gastric cancer and the induction of persistent stomach infections that may lead to peptic ulcers. In recent decades, s...
Profiling of T cell repertoire in peripheral blood of patients from type 2 diabetes with complication
More than 90% of patients with diabetes worldwide are type 2 diabetes (T2D), which is caused by insulin resistance or impaired producing insulin by pancreatic β cells. T2D and its complications, mainly large c...
Tumor microenvironment and immune system preservation in early-stage breast cancer: routes for early recurrence after mastectomy and treatment for lobular and ductal forms of disease
Intra-ductal cancer (IDC) is the most common type of breast cancer, with intra-lobular cancer (ILC) coming in second. Surgery is the primary treatment for early stage breast cancer. There are now irrefutable d...
Predictive biomarkers for immune-related adverse events in cancer patients treated with immune-checkpoint inhibitors
The objective of this study was to identify potential predictors of immune-related adverse events (irAEs) in cancer patients receiving immune checkpoint inhibitor therapy among serum indexes, case data, and li...
Negative prognostic behaviour of PD-L1 expression in tongue and larynx squamous cell carcinoma and its significant predictive power in combination with PD-1 expression on TILs
Biomarkers that can predict outcome will improve the efficacy of treatment for HNSCC patients. In this regard, we retrospectively evaluated the prognostic effect of PD1, PD-L1, and CD45RO in tongue and larynx ...
Whole blood stimulation provides preliminary evidence of altered immune function following SRC
To implement an approach combining whole blood immune stimulation and causal modelling to estimate the impact of sport-related concussion (SRC) on immune function.
IgG antibody response to SARS-CoV-2 infection and its influencing factors in lymphoma patients
The ability of generating effective humoral immune responses to SARS-CoV-2 infection has not been clarified in lymphoma patients. The study aimed to investigate the antibody (Ab) production after SARS-Cov-2 in...
Polarized Th2 cells attenuate high-fat-diet induced obesity through the suppression of lipogenesis
Immune cells, such as macrophages, B cells, neutrophils and T cell subsets, have been implicated in the context of obesity. However, the specific role of Th2 cells in adipose tissue function has remained elusi...
The predictive value of peripheral blood CD4 cells ATP concentration for immune-related adverse events in advanced non-small cell lung cancer patients
Lung cancer with the highest incidence and mortality in the world. Immune checkpoint inhibitors (ICIs), can bring long-term survival benefits to patients, but also can bring immune-related adverse events (irAE...
Evaluation of the TLR3 involvement during Schistosoma japonicum -induced pathology
Despite the functions of TLRs in the parasitic infections have been extensively reported, few studies have addressed the role of TLR3 in the immune response to Schistosoma japonicum infections. The aim of this st...
PRMT2 silencing regulates macrophage polarization through activation of STAT1 or inhibition of STAT6
Macrophages play significant roles in innate immune responses and are heterogeneous cells that can be polarized into M1 or M2 phenotypes. PRMT2 is one of the type I protein arginine methyltransferases involved...
TRPV1 + neurons alter Staphylococcus aureus skin infection outcomes by affecting macrophage polarization and neutrophil recruitment
The interaction between the nervous system and the immune system can affect the outcome of a bacterial infection. Staphylococcus aureus skin infection is a common infectious disease, and elucidating the relations...
Retraction Note: Oral supplementation of diabetic mice with propolis restores the proliferation capacity and chemotaxis of B and T lymphocytes towards CCL21 and CXCL12 by modulating the lipid profile, the pro-inflammatory cytokine levels and oxidative stress
Cd39 identifies a specific cd8 + t cell population in lung adenocarcinoma-related metastatic pleural effusion.
Malignant pleural effusion (MPE), which is a complex microenvironment that contains numerous immune and tumour signals, is common in lung cancer. Gene alterations, such as driver gene mutations, are believed t...
Dissecting cellular states of infiltrating microenvironment cells in melanoma by integrating single-cell and bulk transcriptome analysis
Cellular states of different immune cells can affect the activity of the whole immune microenvironment.
Sec1 regulates intestinal mucosal immunity in a mouse model of inflammatory bowel disease
Inflammatory bowel disease (IBD) is a common immune-mediated condition with its molecular pathogenesis remaining to be fully elucidated. This study aimed to deepen our understanding of the role of FUT2 in human I...
Screening of four lysosome-related genes in sepsis based on RNA sequencing technology
Screening of lysosome-related genes in sepsis patients to provide direction for lysosome-targeted therapy.
Dominant negative biologics normalise the tumour necrosis factor (TNF-α) induced angiogenesis which exploits the Mycobacterium tuberculosis dissemination
Tumor necrosis factor (TNF) is known to promote T cell migration and increase the expression of vascular endothelial growth factor (VEGF) and chemokines. The administration of Xpro-1595, a dominant-negative TN...
Activation dynamics of antigen presenting cells in vivo against Mycobacterium bovis BCG in different immunized route
Control of Tuberculosis (TB) infection is mainly the result of productive teamwork between T-cell populations and antigen presenting cells (APCs). However, APCs activation at the site of initiating cellular im...
Characteristics of circulating immune cells in HBV-related acute-on-chronic liver failure following artificial liver treatment
Liver failure, which is predominantly caused by hepatitis B (HBV) can be improved by an artificial liver support system (ALSS). This study investigated the phenotypic heterogeneity of immunocytes in patients w...
Putative novel outer membrane antigens multi-epitope DNA vaccine candidates identified by Immunoinformatic approaches to control Acinetobacter baumannii
Multi-epitope polypeptide vaccines, a fusion protein, often have a string-of-beads system composed of various specific peptide epitopes, potential adjuvants, and linkers. When choosing the sequence of various ...
Long-term humoral and cellular immunity after primary SARS-CoV-2 infection: a 20-month longitudinal study
SARS-CoV-2 remains a world-wide health issue. SARS-CoV-2-specific immunity is induced upon both infection and vaccination. However, defining the long-term immune trajectory, especially after infection, is limi...
Exploration of biomarkers for systemic lupus erythematosus by machine-learning analysis
In recent years, research on the pathogenesis of systemic lupus erythematosus (SLE) has made great progress. However, the prognosis of the disease remains poor, and high sensitivity and accurate biomarkers are...
Tacrolimus reverses pemphigus vulgaris serum-induced depletion of desmoglein in HaCaT cells via inhibition of heat shock protein 27 phosphorylation
Glucocorticoids are the first-line treatment for Pemphigus vulgaris (PV), but its serious side effects can be life-threatening for PV patients. Tacrolimus (FK506) has been reported to have an adjuvant treatmen...
Increased infiltration of CD4 + T cell in the complement deficient lymphedema model
Lymphedema is an intractable disease that can be caused by injury to lymphatic vessels, such as by surgical treatments for cancer. It can lead to impaired joint mobility in the extremities and reduced quality ...
Vitamin D and biomarkers of inflammation and oxidative stress among pregnant women: a systematic review of observational studies
This systematic review aimed to map the evidence evaluated the relationship between vitamin D and redox and inflammatory status during gestation.
The upregulation of peripheral CD3 - CD56 + CD16 + natural killer cells correlates with Th1/Th2 imbalance in asthma patients during acute upper respiratory viral infections
The aim of this study is to clarify the changes of peripheral CD3 − CD56 + CD16 + NK cells and their correlation with Th1/Th2 immunity profiles in asthma during the phase of acute upper respiratory viral infections (A...
Humoral immune response and changes in peritoneal cell populations in rats immunized against two Leptospira serovars; serovar patoc and serovar pyrogenes
Leptospirosis is a zoonotic disease caused by Leptospira species. Variations in lipopolysaccharide (LPS) structure in Leptospira are known to be associated with the serovar diversity and antigenicity. Development...
Methionine enkephalin(MENK) upregulated memory T cells in anti-influenza response
Novel prophylactic drugs and vaccination strategies for protection against influenza virus should induce specific effector T-cell immune responses in pulmonary airways and peripheral lymphoid organs. Designing...
Transcriptomic analysis identifies CYP27A1 as a diagnostic marker for the prognosis and immunity in lung adenocarcinoma
The association between lipid metabolism disorder and carcinogenesis is well-established, but there is limited research on the connection between lipid metabolism-related genes (LRGs) and lung adenocarcinoma (...
The digestive system and autoimmunity
Digestive autoimmune conditions are a growing challenge to global health. Risk factors associated with autoimmune digestive diseases are complex, including genetic variation, immunological dysfunction, and var...
Bcl-3 regulates T cell function through energy metabolism
Bcl-3 is a member of the IκB protein family and an essential modulator of NF-κB activity. It is well established that Bcl-3 is critical for the normal development, survival and differentiation of adaptive immu...
The development of a highly sensitive and quantitative SARS-CoV-2 rapid antigen test applying newly developed monoclonal antibodies to an automated chemiluminescent flow-through membrane immunoassay device
Rapid and accurate diagnosis of individuals with SARS-CoV-2 infection is an effective way to prevent and control the spread of COVID-19. Although the detection of SARS‐CoV‐2 viral RNA by RT‐qPCR is the gold st...
Immune cell profiles of idiopathic inflammatory myopathy patients expressed anti-aminoacyl tRNA synthetase or anti-melanoma differentiation-associated gene 5 autoantibodies
Patients with idiopathic inflammatory myopathy (IIM) often express a different type of myositis-specific autoantibodies (MSAs), each associated with different clinical symptoms. Understanding the immunopathoge...
Development and validation of a machine learning-based nomogram for predicting HLA-B27 expression
HLA-B27 positivity is normal in patients undergoing rheumatic diseases. The diagnosis of many diseases requires an HLA-B27 examination.
Function and autophagy of monocyte-derived dendritic cells is affected by hepatitis B virus infection
The role of dendritic cells and the autophagy state of dendritic cells in the immune response of hepatitis B virus (HBV) infection was still controversial. In this study, we carefully examined the phenotype, f...
Evaluation of the relationship between serum interleukin-1β levels and expression of inflammasome-related genes in patients with COVID-19
Inflammasomes are a group of molecules that are strongly involved in causing inflammation. This study aimed to evaluate the expression of NLR family pyrin domain containing 1 (NLRP1), NLRP3, and Apoptosis-asso...
Comparison of the effect of autoclaved and non-autoclaved live soil exposure on the mouse immune system
. Lack of exposure to the natural microbial diversity of the environment has been linked to dysregulation of the immune system and numerous noncommunicable diseases, such as allergies and autoimmune disorders....
The effect of serum origin on cytokines induced killer cell expansion and function
Cytokine-induced killer (CIK) cells have shown promising results in adoptive immunotherapy. However, serum may play a determining role in the large-scale expansion of these cells for clinical applications. Acc...
Efficacy and safety of immune checkpoint inhibitors in Proficient Mismatch Repair (pMMR)/ Non-Microsatellite Instability-High (non-MSI-H) metastatic colorectal cancer: a study based on 39 cohorts incorporating 1723 patients
This study was designed to investigate the efficacy and safety of immune checkpoint inhibitors (ICIs)-based therapy in proficient mismatch repair (pMMR)/non-microsatellite instability-high (non-MSI-H) metastat...
Higher plasma interleukin − 6 levels are associated with lung cavitation in drug-resistant tuberculosis
Lung cavitation is associated with heightened TB transmission and poor treatment outcomes. This study aimed to determine the relationship between systemic inflammation and lung cavitation in drug-resistant TB ...
Featured videos
View featured videos from across the BMC-series journals
Important information
Editorial board
For authors
For editorial board members
For reviewers
- Manuscript editing services
Annual Journal Metrics
2022 Citation Impact 3.0 - 2-year Impact Factor 3.6 - 5-year Impact Factor 0.808 - SNIP (Source Normalized Impact per Paper) 0.783 - SJR (SCImago Journal Rank)
2023 Speed 26 days submission to first editorial decision for all manuscripts (Median) 188 days submission to accept (Median)
2023 Usage 671,800 downloads 176 Altmetric mentions
- More about our metrics
- Follow us on Twitter
BMC Immunology
ISSN: 1471-2172
- General enquiries: [email protected]
Advertisement

About The Journal of Immunology
The Journal of Immunology publishes peer-reviewed manuscripts describing novel findings in all areas of experimental immunology, including both basic and clinical studies.
Read More About the Journal
Editor-in-Chief Gail A. Bishop, Ph.D. Full Editorial Board
Seeking EIC Candidates
ImmunoHorizons is accepting applications For its next Editor-in-Chief

Special Collection: Immunology of Zoonotic Diseases and Relevant Animal Models
Special collection curated by Guest Editor Crystal L. Loving, PhD , featuring Brief Reviews on the immunology of zoonotic diseases and relevant animal models
.jpg?versionId=611)
AAI and its journals, The Journal of Immunology and ImmunoHorizons, have become aware that a predatory publisher is sending emails to authors to solicit manuscript submissions. To better inform our valued authors, we compiled a resource on how to spot a predatory publisher. Learn more by visiting the FAQ

AAI Volunteer Portal

Pillars of Immunology
The Journal of Immunology publishes commentaries highlighting scientific reports that were published more than 15 years ago and that have made a major impact on the course of immunological research today. Read published commentaries here .

Don't miss the exciting Brief Review Collection, coming in the January 15, 2024, issue of The Journal of Immunology!

Sign Up for New Issue Alerts
Wondering when the next issues of The JI or ImmunoHorizons are coming out?
Sign up for email alerts so that you don't miss an single issue!
- Current Issue
- Next in The JI
- Collections
Submit a Manuscript
- Instructions for Authors
- About the JI
- Journal Policies
- The JI Editorial Board
- Rights and Permissions
- Follow The JI on Twitter
- Online ISSN 1550-6606
- Print ISSN 0022-1767
Publications
- The Journal of Immunology
- ImmunoHorizons
- AAI Newsletter
- Additional AAI Publications
General Information
- Advertisers
- Subscribers
- Privacy Policy
Journal Services
- Email Alerts
- ImmunoCasts
- Twitter JI
- Cookie Policy
- Accessibility
- Terms & Conditions
- Get Adobe Acrobat Reader
- © 2024 The American Association of Immunologists, Inc.
This Feature Is Available To Subscribers Only
Sign In or Create an Account

Low C4A copy numbers and higher HERV gene insertion contributes to increased risk of SLE, with absence of association with disease phenotype and disease activity
- Christina Mary Mariaselvam
- Gaurav Seth
- Ryad Tamouza
Hormone and reproductive factors and risk of systemic lupus erythematosus: a Mendelian randomized study
- Runyu Chang
- Shate Xiang
- Xinghong Ding
Exploring the causal relationship between Takayasu arteritis and inflammatory bowel disease using Mendelian randomization
- Xiaoli Pang
- Huizhong Yang

Polyethylene glycol and immunology: aspects of allergic reactions and their mechanisms, as well as ways to prevent them in clinical practice
- Maria Zofia Lisiecka

mTOR-mediated differentiation and maintenance of suppressive T cells at the center stage of IPEX treatment
- Rafael Cardoso Maciel Costa Silva
Assessing the steroid-sparing effect of biological agents in randomized controlled trials for lupus: a scoping review
- Savino Sciascia
- Silvia Grazietta Foddai
- Dario Roccatello
Involvement of gut microbiota in multiple sclerosis—review of a new pathophysiological hypothesis and potential treatment target
- Piotr Olejnik
- Kasper Buczma
- Kaja Kasarełło
Correction to: Characteristics of splenic PD-1 + γδT cells in Plasmodium yoelii nigeriensis infection
- Dianhui Chen
- Xingfei Pan
Correction to: The immune regulatory function of B7-H3 in malignancy: spotlight on the IFN-STAT1 axis and regulation of tumor-associated macrophages
- Jong Dae Ji
Monophosphoryl lipid A as a co-adjuvant in methicillin-resistant Staphylococcus aureus vaccine development: improvement of immune responses in a mouse model of infection
- Mehdi Mirshekar
- Setareh Haghighat
- Mohammad Hossein Yazdi
Mass cytometry revealed the circulating immune cell landscape across different Suzuki stages of Moyamoya disease
- Chenglong Liu
- Jizong Zhao
Acute stress transiently activates macrophages and chemokines in cervical lymph nodes
- Akihiro Dohi
- Tadahide Noguchi
- Yoshiyuki Mori

Chronic hyperglycemia impairs anti-microbial function of macrophages in response to Mycobacterium tuberculosis infection
- Gaurav Kumar Chaubey
- Radheshyam Modanwal
Rapid generation of an RBL cellular model to study proteins that cause allergenic reactions in vitro
- Israel Hernández-Aguilar
- Juan Carlos Vizuet-de-Rueda
- Luis M. Teran

Study of pathogenic T-helper cell subsets in Asian Indian patients with Takayasu arteritis
- P. M. Punithavathy
- Ramesh Babu Telugu
- Ruchika Goel

Blood and CSF anti-neuronal antibodies testing in psychotic syndromes: a retrospective analysis from a tertiary psychiatric hospital
- Joana Lopes
- Maria João Malaquias
- Ana Paula Correia
Rituximab alleviates pediatric systemic lupus erythematosus associated refractory immune thrombocytopenia: a case-based review
- Qiaoyan Guo
Evidence of concerning decline of COVID-19 vaccination in older persons
- Camilla Mattiuzzi
- Giuseppe Lippi

The immune regulatory function of B7-H3 in malignancy: spotlight on the IFN-STAT1 axis and regulation of tumor-associated macrophages

Characteristics of splenic PD-1 + γδT cells in Plasmodium yoelii nigeriensis infection

APLAID complicated with arrhythmogenic dilated cardiomyopathy caused by a novel PLCG2 variant
- Tianjiao Wang
- Xiaolin Liu
Evolutionary preservation of CpG dinucleotides in RAG1 may elucidate the relatively high rate of methylation-mediated mutagenesis of RAG1 transposase
- Mariam M. Fawzy
- Maiiada H. Nazmy
- Moustafa Fathy

DNA methylation profiling of labial salivary gland tissues revealed hypomethylation of B-cell-related genes in primary Sjögren’s syndrome
- Jayakanthan Kabeerdoss
- Prabavathi Devarajalu
- Pulukool Sandhya

Response to: regarding the significance of anti-COVID-IgA antibody response in COVID-19 breakthrough infection
- Sabiha Anis
- Mariam Ashfaq Khan
- Samreen Sarfaraz
The combination of IDO and AHR blockers reduces the migration and clonogenicity of breast cancer cells
- Maryam Soltani-asl
- Parviz Azimnasab-sorkhabi
- Jose Roberto Kfoury Jr.
Conventional Tregs in treatment-naïve rheumatoid arthritis are deficient in suppressive function with an increase in percentage of CXCR3 and CCR6 expressing Tregs
- Vallayyachari Kommoju
- Vir Singh Negi
Serum soluble LYVE1 is a promising non-invasive biomarker of renal fibrosis: a population-based retrospective cross-sectional study

COVID-19 in Systemic Lupus Erythematosus patients treated with belimumab: a retrospective clinical study
- Martin Herrmann
Regarding the significance of anti-COVID-IgA antibody response in COVID-19 breakthrough infection
An epicutaneous therapeutic pollen-allergen extract delivery system in an allergic rhinitis mouse model: based on allergen loading on dc-specific aptamers conjugated nanogolds.
- Safoora Pordel
- Navideh Haghnavaz
- Mojtaba Sankian
Discouraging Non-ELISA antiphospholipid antibody assays in antiphospholipid syndrome classification may hinder clinical research
- Xiaochun Susan Zhang
- Nicola Bizzaro
- Jan Damoiseaux
Quantitative evaluation of citrullinated fibrinogen for detection of neutrophil extracellular traps
- Tsubasa Sue
- Tomoki Ichikawa
- Yumiko Higuchi

Long non-coding RNA ENSMUST00000197208 promotes a shift in the Th17/Treg ratio via the P2X7R-NLRP3 inflammasome axis in collagen-induced arthritis

Correction: Major histocompatibility complex class I molecule expression by pancreatic cancer cells is regulated by activation and inhibition of the epidermal growth factor receptor
- Shelby M. Knoche
- Alaina C. Larson
- Joyce C. Solheim
Immune cells and hypertension
A ubiquitin–proteasome system-related signature to predict prognosis, immune infiltration, and therapy efficacy for breast cancer
- Meihuan Wang
- Huawei Zhang

Fructus lycii oligosaccharide alleviates acute liver injury via PI3K/Akt/mTOR pathway
- Xingxing Zhang
- Qingtong Yu

Impact of microparticles released during murine systemic inflammation on macrophage activity and reactive nitrogen species regulation
- Weronika Ortmann
- Elzbieta Kolaczkowska
Comparative analyses of various IgE-mediated and non-IgE-mediated inducers of mast cell degranulation for in vitro study
- Sunisa Yoodee
- Chuda Rujitharanawong
- Visith Thongboonkerd

Semaphorin 3 a restores the ability of type 1 regulatory T cells to suppress food allergy
- Pengyuan Zheng

Insights into the immunological description of cryoglobulins with regard to detection and characterization in Slovenian rheumatological patients
- Manca Ogrič
- Saša Čučnik

Increased synthesis and intestinal expression of IL-39 in patients with inflammatory bowel disease
- Gabriela Fonseca-Camarillo
- Janette Furuzawa-Carballeda
- Jesús K. Yamamoto-Furusho
Role of LINC00240 on T-helper 9 differentiation in allergic rhinitis through influencing DNMT1-dependent methylation of PU.1
- JianGuo Liu
- XunShuo Jiang
- ChunPing Yang

Association of IL-23R and IL-10 variations with Behçet disease: a genetic analysis study
- Guven Yenmis
- Sema Sabancelebi
- Tumay Sadikoglu
Prevalence and sociodemographic correlates of antinuclear antibody testing by indirect immunofluorescence or solid-phase assays in a Spanish population: the Camargo Cohort
- Juan Irure-Ventura
- Daniel Martínez-Revuelta
- José Luis Hernández

Rational design of multi-epitope-based vaccine by exploring all dengue virus serotypes proteome: an immunoinformatic approach
- Ahad Amer Alsaiari
- Mohammed Ageeli Hakami
- Farzana Yasmin

Differential expression of TOB/BTG family members in patients with plaque psoriasis: cross-sectional study
- Carlos A. Barrera-Ochoa

Association study between killer immunoglobulin-like receptor polymorphisms and susceptibility to COVID-19 disease: a systematic review and meta-analysis
- Saeed Hosseini Teshnizi
- Sara Mirzazadeh
- Kurosh Kalantar

Palpitation in women with silicone breast implants: association with autoantibodies against autonomic nervous system
- Gilad Halpert
- Yehuda Shoenfeld
Current genetic defects in common variable immunodeficiency patients on the geography between Europe and Asia: a single-center experience
- Ezgi Topyıldız
- Necil Kutukculer
- Find a journal
- Publish with us
- Track your research
Advertisement

About the Journal
Cancer Immunology Research publishes outstanding original articles reporting major advances in cancer immunology that span the discipline from basic investigations in host-tumor interactions to developmental therapeutics in model systems, early translational studies in patients, and late-stage clinical trials. Read More About the Journal
Editors-in-Chief
Robert D. Schreiber Philip D. Greenberg
Impact Factor
Featured articles.

High-Specificity CRISPR-Mediated Genome Engineering in Anti-BCMA Allogeneic CAR T Cells Suppresses Allograft Rejection in Preclinical Models
Émilie Degagné, et al.

Neutrophils Mediate Protection Against Colitis and Carcinogenesis by Controlling Bacterial Invasion and IL22 Production by γδ T Cells
Silvia Carnevale, et al.

Immune Modulation with RANKL Blockade through Denosumab Treatment in Patients with Cancer
Hewitt Chang, et al.
Trending - Altmetric
The X live feed is not currently active.
Keep up to date with Cancer Immunology Research
Follow @CIR_AACR
- Online First
- Online ISSN 2326-6074
- Print ISSN 2326-6066
AACR Journals
- Blood Cancer Discovery
- Cancer Discovery
- Cancer Epidemiology, Biomarkers & Prevention
- Cancer Immunology Research
- Cancer Prevention Research
- Cancer Research
- Cancer Research Communications
- Clinical Cancer Research
- Molecular Cancer Research
- Molecular Cancer Therapeutics
- Info for Advertisers
- Information for Institutions/Librarians
- Privacy Policy
- Copyright © 2023 by the American Association for Cancer Research.
This Feature Is Available To Subscribers Only
Sign In or Create an Account
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
April 11, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
Research team develops novel PTPN2/N1 inhibitor for cancer immunotherapy using generative AI
by InSilico Medicine
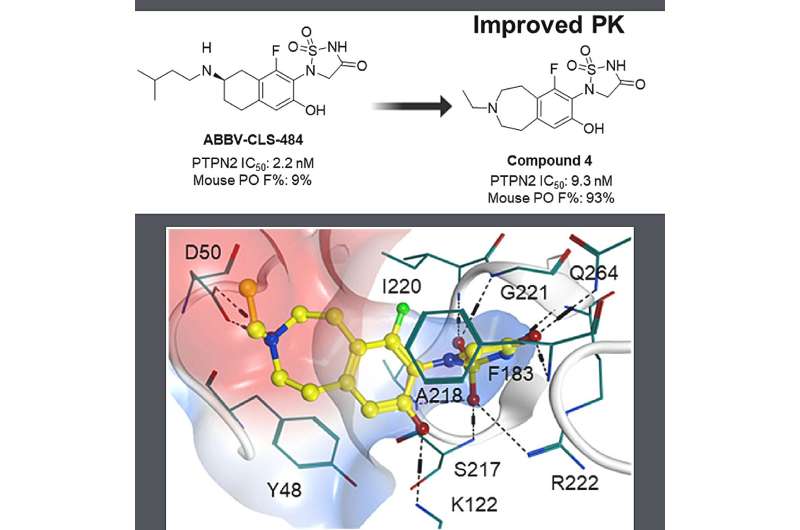
In recent years, cancer immunotherapy, exemplified by PD-1 and its ligand PD-L1 blockade, has made remarkable advances. But while immunotherapy drugs offer new treatment possibilities, only about 20% to 40% of patients respond to these treatments. The majority either don't respond or develop drug resistance. Researchers are now looking for ways to enhance the scope of tumor immunotherapy in order to benefit a wider range of patients.
One such avenue is through the protein tyrosine phosphatase non-receptor type 2 (PTPN2) and its close superfamily member, PTPN1, identified in previous research as crucial modulators involved in the regulation of immune cells signaling pathways that promote tumorigenesis by attenuating tumor-directed immunity. While promising, the development of PTPN2/PTPN1 inhibitors has faced challenges as a result of unfavorable pharmacokinetics due to the highly cationic active site and the relatively shallow nature of the protein surface.
In a significant milestone , researchers at Abbvie discovered the dual PTPN2/N1 inhibitor ABBV-CLS-484 through structure-based drug design and optimization of drug-like properties. Now, clinical stage artificial intelligence (AI)-driven drug discovery company Insilico Medicine ("Insilico") has initiated a program with a fast-follow strategy to design a novel PTPN2/N1 inhibitor with drug-likeness properties and in vivo oral absorption, supported by the Company's generative AI drug design engine Chemistry42 .
The research was published in the European Journal of Medicinal Chemistry .
Scientists entered the structure of the known PTPN2/N1 inhibitor as a reference compound into Chemistry42 as a starting point and generated a series of novel PTPN2/N1 inhibitors based on ligand-based drug design strategy. They further optimized and synthesized the most promising molecules and obtained candidates with desirable ADME properties.
Insilico's compound demonstrated enhanced oral absorption, systemic exposure, and equivalent biological activities compared to the reference compound in in vitro studies. Furthermore, Insilico's compound demonstrated the same efficacious dose as the reference compound in a murine model.
"One of the most significant advances in the research was validating the fast follow ability of Chemistry42, the molecular generation and design engine of Pharma.AI, which allows users to improve existing molecules with more desirable properties rapidly," said Xiao Ding, Ph.D., vice president and head of medicinal chemistry of Insilico Medicine.
"In this paper, we reported a novel PTPN2/PTPN1 inhibitor demonstrating nanomolar inhibitory potency, good in vivo oral bioavailability, and robust in vivo antitumor efficacy. Further investigation is currently ongoing."
Explore further
Feedback to editors
Study reveals potential to reverse lung fibrosis using the body's own healing technique
Researchers discover cell 'crosstalk' that triggers cancer cachexia

Study improves understanding of effects of household air pollution during pregnancy

Wearable sensors for Parkinson's can improve with machine learning, data from healthy adults
2 hours ago

New insights on B cells: Researchers explore building better antibodies and curbing autoimmune diseases

Grieving pet owners comforted by 'supernatural' interactions

Study shows AI improves accuracy of skin cancer diagnoses
Cell's 'garbage disposal' may have another role: Helping neurons near skin sense the environment
Chlamydia vaccine shows promise in early trial

Researchers find no link between COVID-19 virus and development of asthma in children
Related stories.

AI-driven platform discovers PHD inhibitor for anemia treatment
Jan 19, 2024
Using generative AI to identify potent and selective MYT1 inhibitors for the treatment of cancer
Cancer immunotherapy candidate provokes powerful dual response in cancer and immune cells
Oct 4, 2023

Study combines quantum computing and generative AI for drug discovery
May 19, 2023
New study uses AlphaFold and AI to accelerate design of novel drug for liver cancer
Jan 19, 2023

Drug found to correct gene defect that causes immune-driven gut leakiness
Sep 30, 2020
Recommended for you
Scientists uncover a missing link between poor diet and higher cancer risk
5 hours ago

Genomic deletions explain why some types of melanoma resist targeted therapies
3 hours ago
Softer tumors fuel more aggressive spread of triple-negative breast cancer, research shows

Metformin may help the immune system better identify breast cancer cells
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Allergy Asthma Clin Immunol
- v.14(Suppl 2); 2018

An introduction to immunology and immunopathology
Jean s. marshall.
1 Department of Microbiology and Immunology, Dalhousie University, Halifax, NS Canada
Richard Warrington
2 Section of Allergy & Clinical Immunology, Department of Internal Medicine, University of Manitoba, Winnipeg, MB Canada
Wade Watson
3 Division of Allergy, Department of Pediatrics, IWK Health Centre, Dalhousie University, Halifax, NS Canada
Harold L. Kim
4 Western University, London, ON Canada
5 McMaster University, Hamilton, ON Canada
Associated Data
Data sharing not applicable to this article as no datasets were generated or analyzed during the development of this review.
Beyond structural and chemical barriers to pathogens, the immune system has two fundamental lines of defense: innate immunity and adaptive immunity. Innate immunity is the first immunological mechanism for fighting against an intruding pathogen. It is a rapid immune response, initiated within minutes or hours after aggression, that has no immunologic memory. Adaptive immunity, on the other hand, is antigen-dependent and antigen-specific; it has the capacity for memory, which enables the host to mount a more rapid and efficient immune response upon subsequent exposure to the antigen. There is a great deal of synergy between the adaptive immune system and its innate counterpart, and defects in either system can provoke illness or disease, such as inappropriate inflammation, autoimmune diseases, immunodeficiency disorders and hypersensitivity reactions. This article provides a practical overview of innate and adaptive immunity, and describes how these host defense mechanisms are involved in both heath and illness.
There are continuous advances in our current understanding of the immune system and how it functions to protect the body from infection. Given the complex nature of this subject, it is beyond the scope of this article to provide an in-depth review of all aspects of immunology. Rather, the purpose of this article is to provide medical students, medical residents, primary-care practitioners and other healthcare professionals with a basic introduction to the main components and function of the immune system and its role in both health and disease. This article will also serve as a backgrounder to the immunopathological disorders discussed in the remainder of this supplement.
The immune system: innate and adaptive immunity
The immune system refers to a collection of cells, chemicals and processes that function to protect the skin, respiratory passages, intestinal tract and other areas from foreign antigens, such as microbes (organisms such as bacteria, fungi, and parasites), viruses, cancer cells, and toxins. Beyond, the structural and chemical barriers which protect us from infection, the immune system can be simplistically viewed as having two “lines of defense”: innate immunity and adaptive immunity. Innate immunity represents the first line of defense to an intruding pathogen. It is an antigen-independent (non-specific) defense mechanism that is used by the host immediately or within hours of encountering an antigen. The innate immune response has no immunologic memory and, therefore, it is unable to recognize or “memorize” the same pathogen should the body be exposed to it in the future. Adaptive immunity, on the other hand, is antigen-dependent and antigen-specific and, therefore, involves a lag time between exposure to the antigen and maximal response. The hallmark of adaptive immunity is the capacity for memory which enables the host to mount a more rapid and efficient immune response upon subsequent exposure to the antigen. Innate and adaptive immunity are not mutually exclusive mechanisms of host defense, but rather are complementary, with defects in either system resulting in host vulnerability or inappropriate responses [ 1 – 3 ].
Innate immunity
Innate immunity can be viewed as comprising four types of defensive barriers: anatomic (skin and mucous membrane), physiologic (temperature, low pH and chemical mediators), endocytic and phagocytic, and inflammatory. Table 1 summarizes the non-specific host-defense mechanisms for each of these barriers. Cells and processes that are critical for effective innate immunity to pathogens that evade the anatomic barriers have been widely studied. Innate immunity to pathogens relies on pattern recognition receptors (PRRs) which allow a limited range of immune cells to detect and respond rapidly to a wide range of pathogens that share common structures, known as pathogen associated molecular patterns (PAMPs). Examples of these include bacterial cell wall components such as lipopolysaccharides (LPS) and double-stranded ribonucleic acid (RNA) produced during viral infection.
Table 1
Summary of non-specific host-defense mechanisms for barriers of innate immunity [ 1 ]
An important function of innate immunity is the rapid recruitment of immune cells to sites of infection and inflammation through the production of cytokines and chemokines (small proteins involved in cell–cell communication and recruitment). Cytokine production during innate immunity mobilizes many defense mechanisms throughout the body while also activating local cellular responses to infection or injury. Key inflammatory cytokines released during the early response to bacterial infection are: tumour necrosis factor (TNF), interleukin 1 (IL-1) and interleukin 6 (IL-6). These cytokines are critical for initiating cell recruitment and the local inflammation which is essential for clearance of many pathogens. They also contribute to the development of fever. Dysregulated production of such inflammatory cytokines is often associated with inflammatory or autoimmune disease, making them important therapeutic targets.
The complement system is a biochemical cascade that functions to identify and opsonize (coat) bacteria and other pathogens. It renders pathogens susceptible to phagocytosis, a process by which immune cells engulf microbes and remove cell debris, and also kills some pathogens and infected cells directly. The phagocytic action of the innate immune response promotes clearance of dead cells or antibody complexes and removes foreign substances present in organs, tissues, blood and lymph. It can also activate the adaptive immune response through the mobilization and activation of antigen-presenting cells (APCs) (discussed later) [ 1 , 3 ].
Numerous cells are involved in the innate immune response such as phagocytes (macrophages and neutrophils), dendritic cells, mast cells, basophils, eosinophils, natural killer (NK) cells and innate lymphoid cells. Phagocytes are sub-divided into two main cell types: neutrophils and macrophages. Both of these cells share a similar function: to engulf (phagocytose) microbes and kill them through multiple bactericidal pathways. In addition to their phagocytic properties, neutrophils contain granules and enzyme pathways that assist in the elimination of pathogenic microbes. Unlike neutrophils (which are short-lived cells), macrophages are long-lived cells that not only play a role in phagocytosis, but are also involved in antigen presentation to T cells (see Fig. 1 ) [ 1 ].

Characteristics and function of cells involved in innate immunity [ 1 , 3 , 4 ]. *Dust cells (within pulmonary alveolus), histiocytes (connective tissue), Kupffer cells (liver), microglial cells (neural tissue), epithelioid cells (granulomas), osteoclasts (bone), mesangial cells (kidney)
Dendritic cells also phagocytose and function as APCs, initiating the acquired immune response and acting as important messengers between innate and adaptive immunity. Mast cells and basophils share many salient features with each other, and both are instrumental in the initiation of acute inflammatory responses, such as those seen in allergy and asthma. Mast cells also have important functions as immune “sentinel cells” and are early producers of cytokines in response to infection or injury. Unlike mast cells, which generally reside in the connective tissue surrounding blood vessels and are particularly common at mucosal surfaces, basophils reside in the circulation. Eosinophils are granulocytes that possess phagocytic properties and play an important role in the destruction of parasites that are often too large to be phagocytosed. Along with mast cells and basophils, they also control mechanisms associated with allergy and asthma. Natural killer (NK) cells play a major role in the rejection of tumours and the destruction of cells infected by viruses. Destruction of infected cells is achieved through the release of perforins and granzymes (proteins that cause lysis of target cells) from NK-cell granules which induce apoptosis (programmed cell death) [ 4 ]. NK cells are also an important source of another cytokine, interferon-gamma (IFN-γ), which helps to mobilize APCs and promote the development of effective anti-viral immunity. Innate lymphoid cells (ILCs) play a more regulatory role. Depending on their type (i.e., ILC-1, ILC-2, ILC-3), they selectively produce cytokines such as IL-4, IFN-γ and IL-17 that help to direct the appropriate immune response to specific pathogens and contribute to immune regulation in that tissue.
The main characteristics and functions of the cells involved in the innate immune response are summarized in Fig. 1 .
Adaptive immunity
The development of adaptive immunity is aided by the actions of the innate immune system, and is critical when innate immunity is ineffective in eliminating infectious agents. The primary functions of the adaptive immune response are: the recognition of specific “non-self” antigens, distinguishing them from “self” antigens; the generation of pathogen-specific immunologic effector pathways that eliminate specific pathogens or pathogen-infected cells; and the development of an immunologic memory that can quickly eliminate a specific pathogen should subsequent infections occur [ 2 ]. Adaptive immune responses are the basis for effective immunization against infectious diseases. The cells of the adaptive immune system include: antigen-specific T cells, which are activated to proliferate through the action of APCs, and B cells which differentiate into plasma cells to produce antibodies.
T cells and APCs
T cells derive from hematopoietic stem cells in bone marrow and, following migration, mature in the thymus. These cells express a series of unique antigen-binding receptors on their membrane, known as the T-cell receptor (TCR). Each T cell expresses a single type of TCR and has the capacity to rapidly proliferate and differentiate if it receives the appropriate signals. As previously mentioned, T cells require the action of APCs (usually dendritic cells, but also macrophages, B cells, fibroblasts and epithelial cells) to recognize a specific antigen.
The surfaces of APCs express a group of proteins known as the major histocompatibility complex (MHC). MHC are classified as either class I (also termed human leukocyte antigen [HLA] A, B and C) which are found on all nucleated cells, or class II (also termed HLA DP, DQ and DR) which are found only on certain cells of the immune system, including macrophages, dendritic cells and B cells. Class I MHC molecules present endogenous (intracellular) peptides, while class II molecules on APCs present exogenous (extracellular) peptides to T cells. The MHC protein displays fragments of antigens (peptides) when a cell is infected with an intracellular pathogen, such as a virus, or has phagocytosed foreign proteins or organisms [ 2 , 3 ].
T cells have a wide range of unique TCRs which can bind to specific foreign peptides. During the development of the immune system, T cells that would react to antigens normally found in our body are largely eliminated. T cells are activated when they encounter an APC that has digested an antigen and is displaying the correct antigen fragments (peptides) bound to its MHC molecules. The opportunities for the right T cells to be in contact with an APC carrying the appropriate peptide MHC complex are increased by the circulation of T cells throughout the body (via the lymphatic system and blood stream) and their accumulation (together with APCs) in lymph nodes. The MHC-antigen complex activates the TCR and the T cell secretes cytokines which further control the immune response. This antigen presentation process stimulates T cells to differentiate primarily into either cytotoxic T cells (CD8+ cells) or T-helper (Th) cells (CD4+ cells) (see Fig. 2 ). CD8+ cytotoxic T cells are primarily involved in the destruction of cells infected by foreign agents, such as viruses, and the killing of tumour cells expressing appropriate antigens. They are activated by the interaction of their TCR with peptide bound to MHC class I molecules. Clonal expansion of cytotoxic T cells produces effector cells which release substances that induce apoptosis of target cells. Upon resolution of the infection, most effector cells die and are cleared by phagocytes. However, a few of these cells are retained as memory cells that can quickly differentiate into effector cells upon subsequent encounters with the same antigen [ 2 , 3 ].

Adaptive immunity: T-cell and B-cell activation and function. APC antigen-presenting cell, TCR T-cell receptor, MHC major histocompatibility complex
(figure adapted from images available at: http://en.wikipedia.org/wiki/Image:B_cell_activation.png and http://commons.wikimedia.org/wiki/Image:Antigen_presentation.svg )
CD4+ Th cells play an important role in establishing and maximizing the immune response. These cells have no cytotoxic or phagocytic activity, and cannot directly kill infected cells or clear pathogens. However, they “mediate” the immune response by directing other cells to perform these tasks and regulate the type of immune response that develops. Th cells are activated through TCR recognition of antigen bound to class II MHC molecules. Once activated, Th cells release cytokines that influence the activity of many cell types, including the APCs that activate them.
Several types of Th cell responses can be induced by an APC, with Th1, Th2 and Th17 being the most frequent. The Th1 response is characterized by the production of IFN-γ which activates the bactericidal activities of macrophages and enhances anti-viral immunity as well as immunity to other intracellular pathogens. Th1-derived cytokines also contribute to the differentiation of B cells to make opsonizing antibodies that enhance the efficiency of phagocytes. An inappropriate Th1 response is associated with certain autoimmune diseases.
The Th2 response is characterized by the release of cytokines (IL-4, 5 and 13) which are involved in the development of immunoglobulin E (IgE) antibody-producing B cells, as well as the development and recruitment of mast cells and eosinophils that are essential for effective responses against many parasites. In addition, they enhance the production of certain forms of IgG that aid in combatting bacterial infection. As mentioned earlier, mast cells and eosinophils are instrumental in the initiation of acute inflammatory responses, such as those seen in allergy and asthma. IgE antibodies are also associated with allergic reactions (see Table 2 ). Therefore, an imbalance of Th2 cytokine production is associated with the development of atopic (allergic) conditions. Th17 cells have been more recently described. They are characterized by the production of cytokines of the IL-17 family, and are associated with ongoing inflammatory responses, particularly in chronic infection and disease. Like cytotoxic T cells, most Th cells will die upon resolution of infection, with a few remaining as Th memory cells [ 2 , 3 ].
Table 2
Major functions of human Ig antibodies [ 5 ]
A subset of the CD4+ T cell, known as the regulatory T cell (T reg), also plays a role in the immune response. T reg cells limit and suppress immune responses and, thereby, may function to control aberrant responses to self-antigens and the development of autoimmune disease. T reg cells may also help in the resolution of normal immune responses, as pathogens or antigens are eliminated. These cells also play a critical role in the development of “immune tolerance” to certain foreign antigens, such as those found in food.
B cells arise from hematopoietic stem cells in the bone marrow and, following maturation, leave the marrow expressing a unique antigen-binding receptor on their membrane. Unlike T cells, B cells can recognize antigens directly, without the need for APCs, through unique antibodies expressed on their cell surface. The principal function of B cells is the production of antibodies against foreign antigens which requires their further differentiation [ 2 , 3 ]. Under certain circumstances, B cells can also act as APCs.
When activated by foreign antigens to which they have an appropriate antigen specific receptor, B cells undergo proliferation and differentiate into antibody-secreting plasma cells or memory B cells (see Fig. 2 ). Memory B cells are “long-lived” survivors of past infection and continue to express antigen-binding receptors. These cells can be called upon to respond quickly by producing antibodies and eliminating an antigen upon re-exposure. Plasma cells, on the other hand, are relatively short-lived cells that often undergo apoptosis when the inciting agent that induced the immune response is eliminated. However, these cells produce large amounts of antibody that enter the circulation and tissues providing effective protection against pathogens.
Given their function in antibody production, B cells play a major role in the humoral or antibody-mediated immune response (as opposed to the cell-mediated immune response, which is governed primarily by T cells) [ 2 , 3 ].
Antibody-mediated vs. cell-mediated immunity
Antibody-mediated immunity is the branch of the acquired immune system that is mediated by B-cell-antibody production. The antibody-production pathway begins when the B cell’s antigen-binding receptor recognizes and binds to antigen in its native form. Local Th cells secrete cytokines that help the B cell multiply and direct the type of antibody that will be subsequently produced. Some cytokines, such as IL-6, help B-cells to mature into antibody-secreting plasma cells. The secreted antibodies bind to antigens on the surface of pathogens, flagging them for destruction through complement activation, opsonin promotion of phagocytosis and pathogen elimination by immune effector cells. Upon elimination of the pathogen, the antigen–antibody complexes are cleared by the complement cascade (see Fig. 2 ) [ 2 ].
Five major types of antibodies are produced by B cells: IgA, IgD, IgE, IgG and IgM. IgG antibodies can be further subdivided into structurally distinct subclasses with differing abilities to fix complement, act as opsonins, etc. The major classes of antibodies have substantially different biological functions and recognize and neutralize specific pathogens. Table 2 summarizes the various functions of the five Ig antibodies [ 5 ].
Antibodies play an important role in containing virus proliferation during the acute phase of infection. However, they are not generally capable of eliminating a virus once infection has occurred. Once an infection is established, cell-mediated immune mechanisms are most important in host defense against most intracellular pathogens.
Cell-mediated immunity does not involve antibodies, but rather protects an organism through [ 2 ]:
- The activation of antigen-specific cytotoxic T cells that induce apoptosis of cells displaying foreign antigens or derived peptides on their surface, such as virus-infected cells, cells with intracellular bacteria, and cancer cells displaying tumour antigens;
- The activation of macrophages and NK cells, enabling them to destroy intracellular pathogens; and
- The stimulation of cytokine (such as IFNγ) production that further mediates the effective immune response.
Cell-mediated immunity is directed primarily at microbes that survive in phagocytes as well as those that infect non-phagocytic cells. This type of immunity is most effective in eliminating virus-infected cells and cancer cells, but can also participate in defending against fungi, protozoa, cancers, and intracellular bacteria. Cell-mediated immunity also plays a major role in transplant rejection.
Passive vs. active immunization
Acquired immunity is attained through either passive or active immunization. Passive immunization refers to the transfer of active humoral immunity, in the form of “ready-made” antibodies, from one individual to another. It can occur naturally by transplacental transfer of maternal antibodies to the developing fetus, or it can be induced artificially by injecting a recipient with exogenous antibodies that are usually manufactured for this purpose and that are targeted to a specific pathogen or toxin. The latter is used when there is a high risk of infection and insufficient time for the body to develop its own immune response, or to reduce the symptoms of chronic or immunosuppressive diseases.
Active immunization refers to the production of antibodies against a specific antigen or pathogen after exposure to the antigen. It can be acquired through either natural infection with a microbe or through administration of a vaccine that can consist of attenuated (weakened) pathogens, inactivated organisms or specific proteins or carbohydrates known to induce immunity. Effective active immunization often requires the use of “adjuvants” which improve the ability of the immune system to respond to antigen injection.
Immunopathology
As mentioned earlier, defects or malfunctions in either the innate or adaptive immune response can provoke illness or disease. Such disorders are generally caused by an overactive immune response (known as hypersensitivity reactions), an inappropriate reaction to self (known as autoimmunity) or ineffective immune responses (known as immunodeficiency).
Hypersensitivity reactions
Hypersensitivity reactions refer to undesirable responses produced by the normal immune system. There are four types of hypersensitivity reactions [ 6 , 7 ]:
- Type I: immediate hypersensitivity.
- Type II: cytotoxic or antibody-dependent hypersensitivity.
- Type III: immune complex disease.
- Type IV: delayed-type hypersensitivity.
Type I hypersensitivity is the most common type of hypersensitivity reaction. It is an allergic reaction provoked by re-exposure to a specific type of antigen, referred to as an allergen. Unlike the normal immune response, the type I hypersensitivity response is characterized by the secretion of IgE by plasma cells. IgE antibodies bind to receptors on the surface of tissue mast cells and blood basophils, causing them to be “sensitized”. Later exposure to the same allergen cross-links the bound IgE on sensitized cells resulting in degranulation and the secretion of active mediators such as histamine, leukotrienes, and prostaglandins that cause vasodilation and smooth-muscle contraction of the surrounding tissue. Common environmental allergens inducing IgE-mediated allergies include pet (e.g., cat, dog, horse) epithelium, pollen, house dust mites, and molds. Food allergens are also a common cause of type I hypersensitivity reactions, however, these types of reactions are more frequently seen in children than adults. Treatment of type I reactions generally involves trigger avoidance, and in the case of inhaled allergens, pharmacological intervention with bronchodilators, antihistamines and anti-inflammatory agents. Some types of allergic disease can be treated with immunotherapy (see Allergen-specific Immunotherapy article in this supplement). Severe cases of type 1 hypersensitivity (anaphylaxis) may require immediate treatment with epinephrine.
Type II hypersensitivity reactions are rare and take anywhere from 2 to 24 h to develop. These types of reactions occur when IgG and IgM antibodies bind to the patient’s own cell-surface molecules, forming complexes that activate the complement system. This, in turn, leads to opsonization, red blood cell agglutination (process of agglutinating or “clumping together”), cell lysis and death. Some examples of type II hypersensitivity reactions include: erythroblastosis fetalis, Goodpasture syndrome, and autoimmune anemias.
Type III hypersensitivity reactions occur when IgG and IgM antibodies bind to soluble proteins (rather than cell surface molecules as in type II hypersensitivity reactions) forming immune complexes that can deposit in tissues, leading to complement activation, inflammation, neutrophil influx and mast cell degranulation. This type of reaction can take days, or even weeks, to develop and treatment generally involves anti-inflammatory agents and corticosteroids. Examples of type III hypersensitivity reactions include systemic lupus erythematosus (SLE), serum sickness and reactive arthritis.
Unlike the other types of hypersensitivity reactions, type IV reactions are cell-mediated and antibody-independent. They are the second most common type of hypersensitivity reaction and usually take 2 or more days to develop. These types of reactions are caused by the overstimulation of T cells and monocytes/macrophages which leads to the release of cytokines that cause inflammation, cell death and tissue damage. In general, these reactions are easily resolvable through trigger avoidance and the use of topical corticosteroids. An example of this is the skin response to poison ivy.
A brief summary of the four types of hypersensitivity reactions is provided in Table 3 .
Table 3
Types of hypersensitivity reactions [ 6 , 7 ]
Autoimmunity
Autoimmunity involves the loss of normal immune homeostasis such that the organism produces an abnormal response to its own tissue. The hallmark of autoimmunity is the presence of self-reactive T cells, auto-antibodies, and inflammation. Prominent examples of autoimmune diseases include: Celiac disease, type 1 diabetes mellitus, Addison’s disease and Graves’ disease [ 8 ].
Inflammation
Poorly regulated inflammatory responses and tissue damage as a result of inflammation are often immunopathological features. Defects in immune regulation are associated with many chronic inflammatory diseases, including: rheumatoid arthritis, psoriasis, inflammatory bowel disease and asthma. Classical features of inflammation are heat, redness, swelling and pain. Inflammation can be part of the normal host response to infection and a required process to rid the body of pathogens, or it may become uncontrolled and lead to chronic inflammatory disease. The overproduction of inflammatory cytokines (such as TNF, IL-1 and IL-6) as well as the recruitment of inflammatory cells (such as neutrophils and monocytes) through the function of chemokines are important drivers of the inflammatory process. Additional mediators produced by recruited and activated immune cells induce changes in vascular permeability and pain sensitivity.
Immunodeficiency
Immunodeficiency refers to a state in which the immune system’s ability to fight infectious disease is compromised or entirely absent. Immunodeficiency disorders may result from a primary genetic defect (primary immunodeficiency—see Primary Immunodeficiency article in this supplement) which can effect either innate or acquired immune function through inhibition of selected immune cells or pathways, or it may be acquired from a secondary cause (secondary immunodeficiency), such as viral or bacterial infections, malnutrition, autoimmunity or treatment with drugs that induce immunosuppression. Certain diseases can also directly or indirectly impair the immune system such as leukemia and multiple myeloma. Immunodeficiency is also the hallmark of acquired immunodeficiency syndrome (AIDS), caused by the human immunodeficiency virus (HIV). HIV directly infects Th cells and also impairs other immune system responses indirectly [ 9 , 10 ].
Innate immunity is the first immunological, non-specific mechanism for fighting against infections. This immune response is rapid, occurring minutes or hours after aggression and is mediated by numerous cells including phagocytes, mast cells, basophils and eosinophils, as well as the complement system. Adaptive immunity develops in conjunction with innate immunity to eliminate infectious agents; it relies on the tightly regulated interplay between T cells, APCs and B cells. A critical feature of adaptive immunity is the development of immunologic memory or the ability of the system to learn or record its experiences with various pathogens, leading to effective and rapid immune responses upon subsequent exposure to the same or similar pathogens. A brief overview of the defining features of innate and adaptive immunity are presented in Table 4 .
Table 4
Overview of the defining features of innate and adaptive immunity [ 1 ]
There is a great deal of synergy between the adaptive immune system and its innate counterpart, and defects in either system can lead to immunopathological disorders, including autoimmune diseases, immunodeficiencies and hypersensitivity reactions. The remainder of this supplement will focus on the appropriate diagnosis, treatment and management of some of these more prominent disorders, particularly those associated with hypersensitivity reactions.
Declarations
Authors’ contributions All authors wrote and/or edited sections of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to extend special thanks to Dr. Francesca Antonetti whose accredited online course entitled “An Introduction to Immunology” provided the foundation and framework for this article. This informative, entry-level course can be accessed through the Excellence in Medical Education (EXCEMED) website at: https://www.excemed.org .
This article is an update to the article entitled, An Introduction to Immunology and Immunopathology, that originally appeared in the supplement, Practical Guide to Allergy and Immunology in Canada, which was published in Allergy, Asthma & Clinical Immunology in 2011 (available at: https://aacijournal.biomedcentral.com/articles/supplements/volume-7-supplement-1 ).
The authors would like to thank Julie Tasso for her editorial services and assistance in the preparation of this manuscript.
Competing interests
Dr. Jean S. Marshall has no competing interests to disclose. Dr. Richard Warrington is the past president of the Canadian Society of Allergy & Clinical Immunology and Editor-in-Chief of Allergy, Asthma & Clinical Immunology. He has received consulting fees and honoraria from Nycomed, CSL Behring, Talecris, Grifols, Novartis and Shire. Dr. Wade Watson is an associate editor of Allergy, Asthma & Clinical Immunology. Dr. Harold Kim is Vice President of the Canadian Society of Allergy and Clinical Immunology, Past President of the Canadian Network for Respiratory Care, and Co-chief Editor of Allergy, Asthma and Clinical Immunology. He has received consulting fees and honoraria for continuing medical education from AstraZeneca, Aralez, Boehringer Ingelheim, CSL Behring, Kaleo, Merck, Novartis, Pediapharm, Sanofi, Shire and Teva.
Availability of data and materials
Consent for publication.
Not applicable.
Ethics approval and consent to participate
Ethics approval and consent to participate are not applicable to this review article.
Publication of this supplement has been supported by AstraZeneca, Boehringer Ingelheim, CSL Behring Canada Inc., MEDA Pharmaceuticals Ltd., Merck Canada Inc., Pfizer Canada Inc., Shire Pharma Canada ULC, Stallergenes Greer Canada, Takeda Canada, Teva Canada Innovation, Aralez Tribute and Pediapharm.
About this supplement
This article has been published as part of Allergy, Asthma & Clinical Immunology Volume 14 Supplement 2, 2018: Practical guide for allergy and immunology in Canada 2018. The full contents of the supplement are available online at https://aacijournal.biomedcentral.com/articles/supplements/volume-14-supplement-2 .
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19
Keith Sacco, Riccardo Castagnoli, … Luigi D. Notarangelo
In COVID-19, NLRP3 inflammasome genetic variants are associated with critical disease and these effects are partly mediated by the sickness symptom complex: a nomothetic network approach
Michael Maes, Walton Luiz Del Tedesco Junior, … Andréa Name Colado Simão
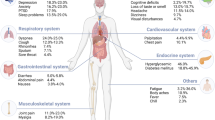
Epidemiology, clinical presentation, pathophysiology, and management of long COVID: an update
Sizhen Su, Yimiao Zhao, … Lin Lu
One in ten severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections results in post-acute sequelae of coronavirus disease 2019 (PASC) or long coronavirus disease (COVID), which affects 65 million people worldwide 1 . Long COVID (LC) remains common, even after mild acute infection with recent variants 2 , and it is likely LC will continue to cause substantial long-term ill health, requiring targeted management based on an understanding of how disease phenotypes relate to underlying mechanisms. Persistent inflammation has been reported in adults with LC 1 , 3 , but studies have been limited in size, timing of samples or breadth of immune mediators measured, leading to inconsistent or absent associations with symptoms. Markers of oxidative stress, metabolic disturbance, vasculoproliferative processes and IFN-, NF-κB- or monocyte-related inflammation have been suggested 3 , 4 , 5 , 6 .
The PHOSP-COVID study, a multicenter United Kingdom study of patients previously hospitalized with COVID-19, has reported inflammatory profiles in 626 adults with health impairment after COVID-19, identified through clustering. Elevated IL-6 and markers of mucosal inflammation were observed in those with severe impairment compared with individuals with milder impairment 7 . However, LC is a heterogeneous condition that may be a distinct form of health impairment after COVID-19, and it remains unclear whether there are inflammatory changes specific to LC symptom subtypes. Determining whether activated inflammatory pathways underlie all cases of LC or if mechanisms differ according to clinical presentation is essential for developing effective therapies and has been highlighted as a top research priority by patients and clinicians 8 .
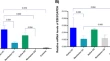
In this Letter, in a prospective multicenter study, we measured 368 plasma proteins in 657 adults previously hospitalized for COVID-19 (Fig. 1a and Table 1 ). Individuals in our cohort experienced a range of acute COVID-19 severities based on World Health Organization (WHO) progression scores 9 ; WHO 3–4 (no oxygen support, n = 133 and median age of 55 years), WHO 5–6 (oxygen support, n = 353 and median age of 59 years) and WHO 7–9 (critical care, n = 171 and median age of 57 years). Participants were hospitalized for COVID-19 ≥3 months before sample collection (median 6.1 months, interquartile range (IQR) 5.1–6.8 months and range 3.0–8.3 months) and confirmed clinically ( n = 36/657) or by PCR ( n = 621/657). Symptom data indicated 233/657 (35%) felt fully recovered at 6 months (hereafter ‘recovered’) and the remaining 424 (65%) reported symptoms consistent with the WHO definition for LC (symptoms ≥3 months post infection 10 ). Given the diversity of LC presentations, patients were grouped according to symptom type (Fig. 1b ). Groups were defined using symptoms and health deficits that have been commonly reported in the literature 1 ( Methods ). A multivariate penalized logistic regression model (PLR) was used to explore associations of clinical covariates and immune mediators at 6 months between recovered patients ( n = 233) and each LC group (cardiorespiratory symptoms, cardioresp, n = 398, Fig. 1c ; fatigue, n = 384, Fig. 1d ; affective symptoms, anxiety/depression, n = 202, Fig. 1e ; gastrointestinal symptoms, GI, n = 132, Fig. 1f ; and cognitive impairment, cognitive, n = 61, Fig. 1g ). Women ( n = 239) were more likely to experience CardioResp (odds ratio (OR 1.14), Fatigue (OR 1.22), GI (OR 1.13) and Cognitive (OR 1.03) outcomes (Fig. 1c,d,f,g ). Repeated cross-validation was used to optimize and assess model performance ( Methods and Extended Data Fig. 1 ). Pre-existing conditions, such as chronic lung disease, neurological disease and cardiovascular disease (Supplementary Table 1 ), were associated with all LC groups (Fig. 1c–g ). Age, C-reactive protein (CRP) and acute disease severity were not associated with any LC group (Table 1 ).
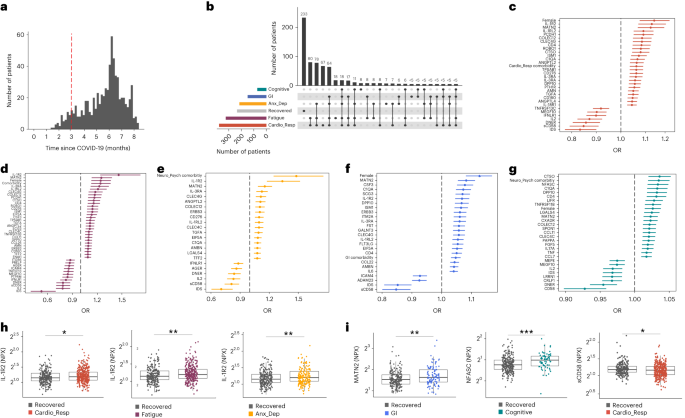
a , Distribution of time from COVID-19 hospitalization at sample collection. All samples were cross-sectional. The vertical red line indicates the 3 month cutoff used to define our final cohort and samples collected before 3 months were excluded. b , An UpSet plot describing pooled LC groups. The horizontal colored bars represent the number of patients in each symptom group: cardiorespiratory (Cardio_Resp), fatigue, cognitive, GI and anxiety/depression (Anx_Dep). Vertical black bars represent the number of patients in each symptom combination group. To prevent patient identification, where less than five patients belong to a combination group, this has been represented as ‘<5’. The recovered group ( n = 233) were used as controls. c – g , Forest plots of Olink protein concentrations (NPX) associated with Cardio_Resp ( n = 365) ( c ), fatigue (n = 314) ( d ), Anx_Dep ( n = 202) ( e ), GI ( n = 124) ( f ) and cognitive ( n = 60) ( g ). Neuro_Psych, neuropsychiatric. The error bars represent the median accuracy of the model. h , i , Distribution of Olink values (NPX) for IL-1R2 ( h ) and MATN2, neurofascin and sCD58 ( i ) measured between symptomatic and recovered individuals in recovered ( n = 233), Cardio_Resp ( n = 365), fatigue ( n = 314) and Anx_Dep ( n = 202) groups ( h ) and MATN2 in GI ( n = 124), neurofascin in cognitive ( n = 60) and sCD58 in Cardio_Resp and recovered groups ( i ). The box plot center line represents the median, the boundaries represent IQR and the whisker length represents 1.5× IQR. The median values were compared between groups using two-sided Wilcoxon signed-rank test, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001.
To study the association of peripheral inflammation with symptoms, we analyzed cross-sectional data collected approximately 6 months after hospitalizations. We measured 368 immune mediators from plasma collected contemporaneously with symptom data. Mediators suggestive of myeloid inflammation were associated with all symptoms (Fig. 1c–h ). Elevated IL-1R2, an IL-1 receptor expressed by monocytes and macrophages modulating inflammation 11 and MATN2, an extracellular matrix protein that modulates tissue inflammation through recruitment of innate immune cells 12 , were associated with cardioresp (IL-1R2 OR 1.14, Fig. 1c,h ), fatigue (IL-1R2 OR 1.45, Fig. 1d,h ), anxiety/depression (IL-1R2 OR 1.34. Fig. 1e,h ) and GI (MATN2 OR 1.08, Fig. 1f ). IL-3RA, an IL-3 receptor, was associated with cardioresp (OR 1.07, Fig. 1c ), fatigue (OR 1.21, Fig. 1d ), anxiety/depression (OR 1.12, Fig. 1e ) and GI (OR 1.06, Fig. 1f ) groups, while CSF3, a cytokine promoting neutrophilic inflammation 13 , was elevated in cardioresp (OR 1.06, Fig. 1c ), fatigue (OR 1.12, Fig. 1d ) and GI (OR 1.08, Fig. 1f ).
Elevated COLEC12, which initiates inflammation in tissues by activating the alternative complement pathway 14 , associated with cardioresp (OR 1.09, Fig. 1c ), fatigue (OR 1.19, Fig. 1d ) and anxiety/depression (OR 1.11, Fig. 1e ), but not with GI (Fig. 1f ) and only weakly with cognitive (OR 1.02, Fig. 1g ). C1QA, a degradation product released by complement activation 15 was associated with GI (OR 1.08, Fig. 1f ) and cognitive (OR 1.03, Fig. 1g ). C1QA, which is known to mediate dementia-related neuroinflammation 16 , had the third strongest association with cognitive (Fig. 1g ). These observations indicated that myeloid inflammation and complement activation were associated with LC.
Increased expression of DPP10 and SCG3 was observed in the GI group compared with recovered (DPP10 OR 1.07 and SCG3 OR 1.08, Fig. 1f ). DPP10 is a membrane protein that modulates tissue inflammation, and increased DPP10 expression is associated with inflammatory bowel disease 17 , 18 , suggesting that GI symptoms may result from enteric inflammation. Elevated SCG3, a multifunctional protein that has been associated with irritable bowel syndrome 19 , suggested that noninflammatory disturbance of the brain–gut axis or dysbiosis, may occur in the GI group. The cognitive group was associated with elevated CTSO (OR 1.04), NFASC (OR 1.03) and SPON-1 (OR 1.02, Fig. 1g,i ). NFASC and SPON-1 regulate neural growth 20 , 21 , while CTSO is a cysteine proteinase supporting tissue turnover 22 . The increased expression of these three proteins as well as C1QA and DPP10 in the cognitive group (Fig. 1g ) suggested neuroinflammation and alterations in nerve tissue repair, possibly resulting in neurodegeneration. Together, our findings indicated that complement activation and myeloid inflammation were common to all LC groups, but subtle differences were observed in the GI and cognitive groups, which may have mechanistic importance. Acutely elevated fibrinogen during hospitalization has been reported to be predictive of LC cognitive deficits 23 . We found elevated fibrinogen in LC relative to recovered (Extended Data Fig. 2a ; P = 0.0077), although this was not significant when restricted to the cognitive group ( P = 0.074), supporting our observation of complement pathway activation in LC and in keeping with reports that complement dysregulation and thrombosis drive severe COVID-19 (ref. 24 ).
Elevated sCD58 was associated with lower odds of all LC symptoms and was most pronounced in cardioresp (OR 0.85, Fig. 1c,i ), fatigue (OR 0.80, Fig. 1d ) and anxiety/depression (OR 0.83, Fig. 1e ). IL-2 was negatively associated with the cardioresp (Fig. 1c , OR 0.87), fatigue (Fig. 1d , OR 0.80), anxiety/depression (Fig. 1e , OR 0.84) and cognitive (Fig. 1g , OR 0.96) groups. Both IL-2 and sCD58 have immunoregulatory functions 25 , 26 . Specifically, sCD58 suppresses IL-1- or IL-6-dependent interactions between CD2 + monocytes and CD58 + T or natural killer cells 26 . The association of sCD58 with recovered suggests a central role of dysregulated myeloid inflammation in LC. Elevated markers of tissue repair, IDS and DNER 27 , 28 , were also associated with recovered relative to all LC groups (Fig. 1c–g ). Taken together, our data suggest that suppression of myeloid inflammation and enhanced tissue repair were associated with recovered, supporting the use of immunomodulatory agents in therapeutic trials 29 (Supplementary Table 2 ).
We next sought to validate the experimental and analytical approaches used. Although Olink has been validated against other immunoassay platforms, showing superior sensitivity and specificity 30 , 31 , we confirmed the performance of Olink against chemiluminescent immunoassays within our cohort. We performed chemiluminescent immunoassays on plasma from a subgroup of 58 participants (recovered n = 13 and LC n = 45). There were good correlations between results from Olink (normalized protein expression (NPX)) and chemiluminescent immunoassays (pg ml −1 ) for CSF3, IL-1R2, IL-3RA, TNF and TFF2 (Extended Data Fig. 3 ). Most samples did not have concentrations of IL-2 detectable using a mesoscale discovery chemiluminescent assay, limiting this analysis to 14 samples (recovered n = 4, LC n = 10, R = 0.55 and P = 0.053, Extended Data Fig. 3 ). We next repeated our analysis using alternative definitions of LC. The Centers for Disease Control and Prevention and National Institute for Health and Care Excellence definitions for LC include symptoms occurring 1 month post infection 32 , 33 . Using the 1 month post-infection definition included 62 additional participants to our analysis (recovered n = 21, 3 females and median age 61 years and LC n = 41, 15 females and median age 60 years, Extended Data Fig. 2c ) and found that inflammatory associations with each LC group were consistent with our analysis based on the WHO definition (Extended Data Fig. 2d–h ). Finally, to validate the analytical approach (PLR) we examined the distribution of data, prioritizing proteins that were most strongly associated with each LC/recovered group (IL-1R2, MATN2, NFASC and sCD58). Each protein was significantly elevated in the LC group compared with recovered (Fig. 1h,i and Extended Data Fig. 4 ), consistent with the PLR. Alternative regression approaches (unadjusted regression models and partial least squares, PLS) reported results consistent with the original analysis of protein associations and LC outcome in the WHO-defined cohort (Fig. 1c–g , Supplementary Table 3 and Extended Data Figs. 5 and 6 ). The standard errors of PLS estimates were wide (Extended Data Fig. 6 ), consistent with previous demonstrations that PLR is the optimal method to analyze high-dimensional data where variables may have combined effects 34 . As inflammatory proteins are often colinear, working in-tandem to mediate effects, we prioritized PLR results to draw conclusions.
To explore the relationship between inflammatory mediators associated with different LC symptoms, we performed a network analysis of Olink mediators highlighted by PLR within each LC group. COLEC12 and markers of endothelial and mucosal inflammation (MATN2, PCDH1, ROBO1, ISM1, ANGPTL2, TGF-α and TFF2) were highly correlated within the cardioresp, fatigue and anxiety/depression groups (Fig. 2 and Extended Data Fig. 7 ). Elevated PCDH1, an adhesion protein modulating airway inflammation 35 , was highly correlated with other inflammatory proteins associated with the cardioresp group (Fig. 2 ), suggesting that systemic inflammation may arise from the lung in these individuals. This was supported by increased expression of IL-3RA, which regulates innate immune responses in the lung through interactions with circulating IL-3 (ref. 36 ), in fatigue (Figs. 1d and 2 ), which correlated with markers of tissue inflammation, including PCDH1 (Fig. 2 ). MATN2 and ISM1, mucosal proteins that enhance inflammation 37 , 38 , were highly correlated in the GI group (Fig. 2 ), highlighting the role of tissue-specific inflammation in different LC groups. SCG3 correlated less closely with mediators in the GI group (Fig. 2 ), suggesting that the brain–gut axis may contribute separately to some GI symptoms. SPON-1, which regulates neural growth 21 , was the most highly correlated mediator in the cognitive group (Fig. 2 and Extended Data Fig. 7 ), highlighting that processes within nerve tissue may underlie this group. These observations suggested that inflammation might arise from mucosal tissues and that additional mechanisms may contribute to pathophysiology underlying the GI and cognitive groups.
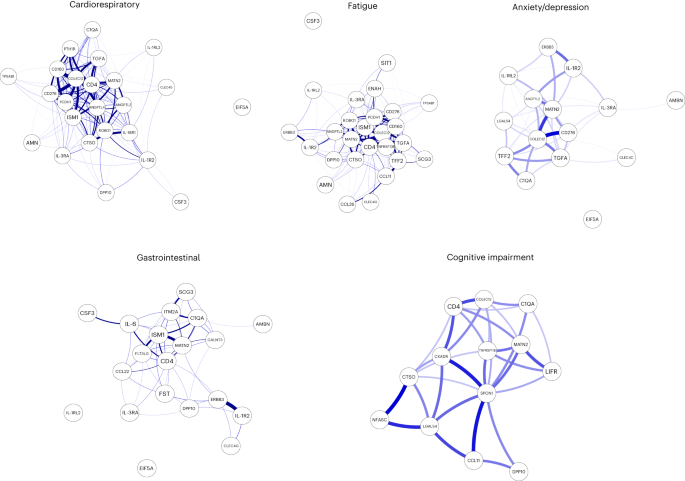
Network analysis of Olink mediators associated with cardioresp ( n = 365), fatigue ( n = 314), anxiety/depression ( n = 202), GI ( n = 124) and cognitive groups ( n = 60). Each node corresponds to a protein mediator identified by PLR. The edges (blue lines) were weighted according to the size of Spearman’s rank correlation coefficient between proteins. All edges represent positive and significant correlations ( P < 0.05) after FDR adjustment.
Women were more likely to experience LC (Table 1 ), as found in previous studies 1 . As estrogen can influence immunological responses 39 , we investigated whether hormonal differences between men and women with LC in our cohort explained this trend. We grouped men and women with LC symptoms into two age groups (those younger than 50 years and those 50 years and older, using age as a proxy for menopause status in women) and compared mediator levels between men and women in each age group, prioritizing those identified by PLR to be higher in LC compared with recovered. As we aimed to understand whether women with LC had stronger inflammatory responses than men with LC, we did not assess differences in men and women in the recovered group. IL-1R2 and MATN2 were significantly higher in women ≥50 years than men ≥50 years in the cardioresp group (Fig. 3a , IL-1R2 and MATN2) and the fatigue group (Fig. 3b ). In the GI group, CSF3 was higher in women ≥50 years compared with men ≥50 years (Fig. 3c ), indicating that the inflammatory markers observed in women were not likely to be estrogen-dependent. Women have been reported to have stronger innate immune responses to infection and to be at greater risk of autoimmunity 39 , possibly explaining why some women in the ≥50 years group had higher inflammatory proteins than men the same group. Proteins associated with the anxiety/depression (IL-1R2 P = 0.11 and MATN2 P = 0.61, Extended Data Fig. 8a ) and cognitive groups (CTSO P = 0.64 and NFASC P = 0.41, Extended Data Fig. 8b ) were not different between men and women in either age group, consistent with the absent/weak association between sex and these outcomes identified by PLR (Fig. 1e,g ). Though our findings suggested that nonhormonal differences in inflammatory responses may explain why some women are more likely to have LC, they require confirmation in adequately powered studies.
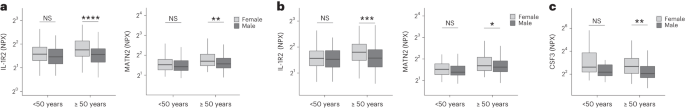
a – c , Olink-measured plasma protein levels (NPX) of IL-1R2 and MATN2 ( a and b ) and CSF3 ( c ) between LC men and LC women divided by age (<50 or ≥50 years) in the cardiorespiratory group (<50 years n = 8 and ≥50 years n = 270) ( a ), fatigue group (<50 years n = 81 and ≥50 years n = 227) ( b ) and GI group (<50 years n = 34 and ≥50 years n = 82) ( c ). the median values were compared between men and women using two-sided Wilcoxon signed-rank test, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001. The box plot center line represents the median, the boundaries represent IQR and the whisker length represents 1.5× IQR.
To test whether local respiratory tract inflammation persisted after COVID-19, we compared nasosorption samples from 89 participants (recovered, n = 31; LC, n = 33; and healthy SARS-CoV-2 naive controls, n = 25, Supplementary Tables 4 and 5 ). Several inflammatory markers were elevated in the upper respiratory tract post COVID (including IL-1α, CXCL10, CXCL11, TNF, VEGF and TFF2) when compared with naive controls, but similar between recovered and LC (Fig. 4a ). In the cardioresp group ( n = 29), inflammatory mediators elevated in plasma (for example, IL-6, APO-2, TGF-α and TFF2) were not elevated in the upper respiratory tract (Extended Data Fig. 9a ) and there was no correlation between plasma and nasal mediator levels (Extended Data Fig. 9b ). This exploratory analysis suggested upper respiratory tract inflammation post COVID was not specifically associated with cardiorespiratory symptoms.
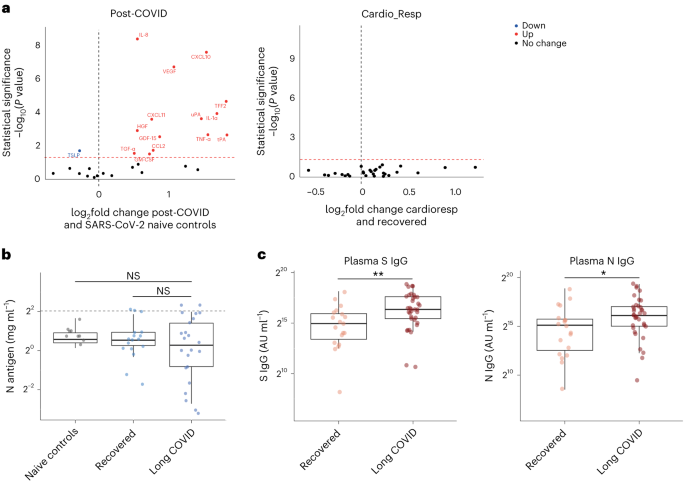
a , Nasal cytokines measured by immunoassay in post-COVID participants ( n = 64) compared with healthy SARS-CoV-2 naive controls ( n = 25), and between the the cardioresp group ( n = 29) and the recovered group ( n = 31). The red values indicate significantly increased cytokine levels after FDR adjustment ( P < 0.05) using two-tailed Wilcoxon signed-rank test. b , SARS-CoV-2 N antigen measured in sputum by electrochemiluminescence from recovered ( n = 17) and pooled LC ( n = 23) groups, compared with BALF from SARS-CoV-2 naive controls ( n = 9). The horizontal dashed line indicates the lower limit of detection of the assay. c , Plasma S- and N-specific IgG responses measured by electrochemiluminescence in the LC ( n = 35) and recovered ( n = 19) groups. The median values were compared using two-sided Wilcoxon signed-rank tests, NS P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001. The box plot center lines represent the median, the boundaries represent IQR and the whisker length represents 1.5× IQR.
To explore whether SARS-CoV-2 persistence might explain the inflammatory profiles observed in the cardioresp group, we measured SARS-CoV-2 nucleocapsid (N) antigen in sputum from 40 participants (recovered n = 17 and LC n = 23) collected approximately 6 months post hospitalization (Supplementary Table 6 ). All samples were compared with prepandemic bronchoalveolar lavage fluid ( n = 9, Supplementary Table 4 ). Only four samples (recovered n = 2 and LC n = 2) had N antigen above the assay’s lower limit of detection, and there was no difference in N antigen concentrations between LC and recovered (Fig. 4b , P = 0.78). These observations did not exclude viral persistence, which might require tissues samples for detection 40 , 41 . On the basis of the hypothesis that persistent viral antigen might prevent a decline in antibody levels over time, we examined the titers of SARS-CoV-2-specific antibodies in unvaccinated individuals (recovered n = 19 and LC n = 35). SARS-CoV-2 N-specific ( P = 0.023) and spike (S)-specific ( P = 0.0040) immunoglobulin G (IgG) levels were elevated in LC compared with recovered (Fig. 4c ).
Overall, we identified myeloid inflammation and complement activation in the cardioresp, fatigue, anxiety/depression, cognitive and GI groups 6 months after hospitalization (Extended Data Fig. 10 ). Our findings build on results of smaller studies 5 , 6 , 42 and are consistent with a genome-wide association study that identified an independent association between LC and FOXP4 , which modulates neutrophilic inflammation and immune cell function 43 , 44 . In addition, we identified tissue-specific inflammatory elements, indicating that myeloid disturbance in different tissues may result in distinct symptoms. Multiple mechanisms for LC have been suggested, including autoimmunity, thrombosis, vascular dysfunction, SARS-CoV-2 persistence and latent virus reactivation 1 . All these processes involve myeloid inflammation and complement activation 45 . Complement activation in LC has been suggested in a proteomic study in 97 mostly nonhospitalized COVID-19 cases 42 and a study of 48 LC patients, of which one-third experienced severe acute disease 46 . As components of the complement system are known to have a short half-life 47 , ongoing complement activation suggests active inflammation rather than past tissue damage from acute infection.
Despite the heterogeneity of LC and the likelihood of coexisting or multiple etiologies, our work suggests some common pathways that might be targeted therapeutically and supports the rationale for several drugs currently under trial. Our finding of increased sCD58 levels (associated with suppression of monocyte–lymphocyte interactions 26 ) in the recovered group, strengthens our conclusion that myeloid inflammation is central to the biology of LC and that trials of steroids, IL-1 antagonists, JAK inhibitors, naltrexone and colchicine are justified. Although anticoagulants such as apixaban might prevent thrombosis downstream of complement dysregulation, they can also increase the risk of serious bleeding when given after COVID-19 hospitalization 48 . Thus, clinical trials, already underway, need to carefully assess the risks and benefits of anticoagulants (Supplementary Table 2 ).
Our finding of elevated S- and N-specific IgG in LC could suggest viral persistence, as found in other studies 6 , 42 , 49 . Our network analysis indicated that inflammatory proteins in the cardioresp group interacted strongly with ISM1 and ROBO1, which are expressed during respiratory tract infection and regulate lung inflammation 50 , 51 . Although we were unable to find SARS-CoV-2 antigen in sputum from our LC cases, we did not test for viral persistence in GI tract and lung tissue 40 , 41 or in plasma 52 . Evidence of SARS-CoV-2 persistence would justify trials of antiviral drugs (singly or in combination) in LC. It is also possible that autoimmune processes could result in an innate inflammatory profile in LC. Autoreactive B cells have been identified in LC patients with higher SARS-CoV-2-specific antibody titers in a study of mostly mild acute COVID cases (59% WHO 2–3) 42 , a different population from our study of hospitalized cases.
Our observations of distinct protein profiles in GI and cognitive groups support previous reports on distinct associations between Epstein–Barr virus reactivation and neurological symptoms, or autoantibodies and GI symptoms relative to other forms of LC 49 , 53 . We did not assess autoantibody induction but found evidence of brain–gut axis disturbance (SCG3) in the GI group, which occurs in many autoimmune diseases 54 . We found signatures suggestive of neuroinflammation (C1QA) in the cognitive group, consistent with findings of brain abnormalities on magnetic resonance imaging after COVID-19 hospitalization 55 , as well as findings of microglial activation in mice after COVID-19 (ref. 56 ). Proinflammatory signatures dominated in the cardioresp, fatigue and anxiety/depression groups and were consistent with those seen in non-COVID depression, suggesting shared mechanisms 57 . The association between markers of myeloid inflammation, including IL-3RA, and symptoms was greatest for fatigue. Whilst membrane-bound IL-3RA facilitates IL-3 signaling upstream of myelopoesis 36 its soluble form (measured in plasma) can bind IL-3 and can act as a decoy receptor, preventing monocyte maturation and enhancing immunopathology 58 . Monocytes from individuals with post-COVID fatigue are reported to have abnormal expression profiles (including reduced CXCR2), suggestive of altered maturation and migration 5 , 59 . Lung-specific inflammation was suggested by the association between PCDH1 (an airway epithelial adhesion molecule 35 ) and cardioresp symptoms.
Our observations do not align with all published observations on LC. One proteomic study of 55 LC cases after generally mild (WHO 2–3) acute disease found that TNF and IFN signatures were elevated in LC 3 . Vasculoproliferative processes and metabolic disturbance have been reported in LC 4 , 60 , but these studies used uninfected healthy individuals for comparison and cannot distinguish between LC-specific phenomena and residual post-COVID inflammation. A study of 63 adults (LC, n = 50 and recovered, n = 13) reported no association between immune cell activation and LC 3 months after infection 61 , though myeloid inflammation was not directly measured, and 3 months post infection may be too early to detect subtle differences between LC and recovered cases due to residual acute inflammation.
Our study has limitations. We designed the study to identify inflammatory markers identifying pathways underlying LC subgroups rather than diagnostic biomarkers. The ORs we report are small, but associations were consistent across alternative methods of analysis and when using different LC definitions. Small effect sizes can be expected when using PLR, which shrinks correlated mediator coefficients to reflect combined effects and prevent colinear inflation 62 , and could also result from measurement of plasma mediators that may underestimate tissue inflammation. Although our LC cohort is large compared with most other published studies, some of our subgroups are small (only 60 cases were designated cognitive). Though the performance of the cognitive PLR model was adequate, our findings should be validated in larger studies. It should be noted that our cohort of hospitalized cases may not represent all types of LC, especially those occurring after mild infection. We looked for an effect of acute disease severity within our study and did not find it, and are reassured that the inflammatory profiles we observed were consistent with those seen in smaller studies including nonhospitalized cases 42 , 46 . Studies of posthospital LC may be confounded by ‘posthospital syndrome’, which encompasses general and nonspecific effects of hospitalization (particularly intensive care) 63 .
In conclusion, we found markers of myeloid inflammation and complement activation in our large prospective posthospital cohort of patients with LC, in addition to distinct inflammatory patterns in patients with cognitive impairment or gastrointestinal symptoms. These findings show the need to consider subphenotypes in managing patients with LC and support the use of antiviral or immunomodulatory agents in controlled therapeutic trials.
Study design and ethics
After hospitalization for COVID-19, adults who had no comorbidity resulting in a prognosis of less than 6 months were recruited to the PHOSP-COVID study ( n = 719). Patients hospitalized between February 2020 and January 2021 were recruited. Both sexes were recruited and gender was self-reported (female, n = 257 and male, n = 462). Written informed consent was obtained from all patients. Ethical approvals for the PHOSP-COVID study were given by Leeds West Research Ethics Committee (20/YH/0225).
Symptom data and samples were prospectively collected from individuals approximately 6 months (IQR 5.1–6.8 months and range 3.0–8.3 months) post hospitalization (Fig. 1a ), via the PHOSP-COVID multicenter United Kingdom study 64 . Data relating to patient demographics and acute admission were collected via the International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization Clinical Characterisation Protocol United Kingdom (ISARIC4C study; IRAS260007/IRAS126600) (ref. 65 ). Adults hospitalized during the SARS-CoV-2 pandemic were systematically recruited into ISARIC4C. Written informed consent was obtained from all patients. Ethical approval was given by the South Central–Oxford C Research Ethics Committee in England (reference 13:/SC/0149), Scotland A Research Ethics Committee (20/SS/0028) and WHO Ethics Review Committee (RPC571 and RPC572l, 25 April 2013).
Data were collected to account for variables affecting symptom outcome, via hospital records and self-reporting. Acute disease severity was classified according to the WHO clinical progression score: WHO class 3–4: no oxygen therapy; class 5: oxygen therapy; class 6: noninvasive ventilation or high-flow nasal oxygen; and class 7–9: managed in critical care 9 . Clinical data were used to place patients into six categories: ‘recovered’, ‘GI’, ‘cardiorespiratory’, ‘fatigue’, ‘cognitive impairment’ and ‘anxiety/depression’ (Supplementary Table 7 ). Patient-reported symptoms and validated clinical scores were used when feasible, including Medical Research Council (MRC) breathlessness score, dyspnea-12 score, Functional Assessment of Chronic Illness Therapy (FACIT) score, Patient Health Questionnaire (PHQ)-9 and Generalized Anxiety Disorder (GAD)-7. Cognitive impairment was defined as a Montreal Cognitive Assessment score <26. GI symptoms were defined as answering ‘Yes’ to the presence of at least two of the listed symptoms. ‘Recovered’ was defined by self-reporting. Patients were placed in multiple groups if they experienced a combination of symptoms.
Matched nasal fluid and sputum samples were prospectively collected from a subgroup of convalescent patients approximately 6 months after hospitalization via the PHOSP-COVID study. Nasal and bronchoalveolar lavage fluid (BALF) collected from healthy volunteers before the COVID-19 pandemic were used as controls (Supplementary Table 4 ). Written consent was obtained for all individuals and ethical approvals were given by London–Harrow Research Ethics Committee (13/LO/1899) for the collection of nasal samples and the Health Research Authority London–Fulham Research Ethics Committee (IRAS project ID 154109; references 14/LO/1023, 10/H0711/94 and 11/LO/1826) for BALF samples.
Ethylenediaminetetraacetic acid plasma was collected from whole blood taken by venepuncture and frozen at −80 °C as previously described 7 , 66 . Nasal fluid was collected using a NasosorptionTM FX·I device (Hunt Developments), which uses a synthetic absorptive matrix to collect concentrated nasal fluid. Samples were eluted and stored as previously described 67 . Sputum samples were collected via passive expectoration and frozen at −80 °C without the addition of buffers. Sputum samples from convalescent individuals were compared with BALF from healthy SARS-CoV-2-naive controls, collected before the pandemic. BALF samples were used to act as a comparison for lower respiratory tract samples since passively expectorated sputum from healthy SARS-CoV-2-naive individuals was not available. BALF samples were obtained by instillation and recovery of up to 240 ml of normal saline via a fiberoptic bronchoscope. BALF was filtered through 100 µM strainers into sterile 50 ml Falcon tubes, then centrifuged for 10 min at 400 g at 4 °C. The resulting supernatant was transferred into sterile 50 ml Falcon tubes and frozen at −80 °C until use. The full methods for BALF collection and processing have been described previously 68 , 69 .
Immunoassays
To determine inflammatory signatures that associated with symptom outcomes, plasma samples were analyzed on an Olink Explore 384 Inflammation panel 70 . Supplementary Table 8 (Appendix 1 ) lists all the analytes measured. To ensure the validity of results, samples were run in a single batch with the use of negative controls, plate controls in triplicate and repeated measurement of patient samples between plates in duplicate. Samples were randomized between plates according to site and sample collection date. Randomization between plates was blind to LC/recovered outcome. Data were first normalized to an internal extension control that was included in each sample well. Plates were standardized by normalizing to interplate controls, run in triplicate on each plate. Each plate contained a minimum of four patient samples, which were duplicates on another plate; these duplicate pairs allowed any plate to be linked to any other through the duplicates. Data were then intensity normalized across all cohort samples. Finally, Olink results underwent quality control processing and samples or analytes that did not reach quality control standards were excluded. Final normalized relative protein quantities were reported as log 2 NPX values.
To further validate our findings, we performed conventional electrochemiluminescence (ECL) assays and enzyme-linked immunosorbent assay for Olink mediators that were associated with symptom outcome ( Supplementary Methods ). Contemporaneously collected plasma samples were available from 58 individuals. Like most omics platforms, Olink measures relative quantities, so perfect agreement with conventional assays that measure absolute concentrations is not expected.
Sputum samples were thawed before analysis and sputum plugs were extracted with the addition of 0.1% dithiothreitol creating a one in two sample dilution, as previously described 71 . SARS-CoV-2 S and N proteins were measured by ECL S-plex assay at a fixed dilution of one in two (Mesoscale Diagnostics), as per the manufacturers protocol 72 . Control BALF samples were thawed and measured on the same plate, neat. The S-plex assay is highly sensitive in detecting viral antigen in respiratory tract samples 73 .
Nasal cytokines were measured by ECL (mesoscale discovery) and Luminex bead multiplex assays (Biotechne). The full methods and list of analytes are detailed in Supplementary Methods .
Statistics and reproducibility
Clinical data was collected via the PHOSP REDCap database, to which access is available under reasonable request as per the data sharing statement in the manuscript. All analyses were performed within the Outbreak Data Analysis Platform (ODAP). All data and code can be accessed using information in the ‘Data sharing’ and ‘Code sharing’ statements at the end of the manuscript. No statistical method was used to predetermine sample size. Data distribution was assumed to be normal but this was not formally tested. Olink assays and immunoassays were randomized and investigators were blinded to outcomes.
To determine protein signatures that associated with each symptom outcome, a ridge PLR was used. PLR shrinks coefficients to account for combined effects within high-dimensional data, preventing false discovery while managing multicollinearity 34 . Thus, PLR was chosen a priori as the most appropriate model to assess associations between a large number of explanatory variables (that may work together to mediate effects) and symptom outcome 34 , 62 , 70 , 74 . In keeping with our aim to perform an unbiased exploration of inflammatory process, the model alpha was set to zero, facilitating regularization without complete penalization of any mediator. This enabled review of all possible mediators that might associate with LC 62 .
A 50 repeats tenfold nested cross-validation was used to select the optimal lambda for each model and assess its accuracy (Extended Data Fig. 1 ). The performance of the cognitive impairment model was influenced by the imbalance in size of the symptom group ( n = 60) relative to recovered ( n = 250). The model was weighted to account for this imbalance resulting in a sensitivity of 0.98, indicating its validity. We have expanded on the model performance and validation approaches in Supplementary Information .
Age, sex, acute disease severity and preexisting comorbidities were included as covariates in the PLR analysis (Supplementary Tables 1 and 3 ). Covariates were selected a priori using features reported to influence the risk of LC and inflammatory responses 1 , 39 , 64 , 75 . Ethnicity was not included since it has been shown not to predict symptom outcome in this cohort 64 . Individuals with missing data were excluded from the regression analysis. Each symptom group was compared with the ‘recovered’ group. The model coefficients of each covariate were converted into ORs for each outcome and visualized in a forest plot, after removing variables associated with regularized OR between 0.98 and 1.02 or in cases where most variables fell outside of this range, using mediators associated with the highest decile of coefficients either side of this range. This enabled exclusion of mediators with effect sizes that were unlikely to have clinical or mechanistic importance since the ridge PLR shrinks and orders coefficients according to their relative importance rather than making estimates with standard error. Thus, confidence intervals cannot be appropriately derived from PLR, and forest plot error bars were calculated using the median accuracy of the model generated by the nested cross-validation. To verify observations made through PLR analysis, we also performed an unadjusted PLR, an unadjusted logistic regression and a PLS analysis. Univariate analyses using Wilcoxon signed-rank test was also performed (Supplementary Table 8 , Appendix 1 ). Analyses were performed in R version 4.2.0 using ‘data.table v1.14.2’, ‘EnvStats v2.7.0’ ‘tidyverse v1.3.2’, ‘lme4 v1.1-32’, ‘caret v6.0-93’, ‘glmnet v4.1-6’, ‘mdatools v0.14.0’, ‘ggpubbr v0.4.0’ and ‘ggplot2 v3.3.6’ packages.
To further investigate the relationship between proteins elevated in each symptom group, we performed a correlation network analysis using Spearman’s rank correlation coefficient and false discovery rate (FDR) thresholding. The mediators visualized in the PLR forest plots, which were associated with cardiorespiratory symptoms, fatigue, anxiety/depression GI symptoms and cognitive impairment were used, respectively. Analyses were performed in R version 4.2.0 using ‘bootnet v1.5.6 ’ and ‘qgraph v1.9.8 ’ packages.
To determine whether differences in protein levels between men and women related to hormonal differences, we divided each symptom group into premenopausal and postmenopausal groups using an age cutoff of 50 years old. Differences between sexes in each group were determined using the Wilcoxon signed-rank test. To understand whether antigen persistence contributed to inflammation in adults with LC, the median viral antigen concentration from sputum/BALF samples and cytokine concentrations from nasal samples were compared using the Wilcoxon signed-rank test. All tests were two-tailed and statistical significance was defined as a P value < 0.05 after adjustment for FDR ( q -value of 0.05). Analyses were performed in R version 4.2.0 using ‘bootnet v1.5.6’ and ‘qgraph v1.9.8’ packages.
Extended Data Fig. 10 was made using Biorender, accessed at www.biorender.com .
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This is an open access article under the CC BY 4.0 license.
The PHOSP-COVID protocol, consent form, definition and derivation of clinical characteristics and outcomes, training materials, regulatory documents, information about requests for data access, and other relevant study materials are available online at ref. 76 . Access to these materials can be granted by contacting [email protected] and [email protected].
The ISARIC4C protocol, data sharing and publication policy are available at https://isaric4c.net . ISARIC4C’s Independent Data and Material Access Committee welcomes applications for access to data and materials ( https://isaric4c.net ).
The datasets used in the study contain extensive clinical information at an individual level that prevent them from being deposited in an public depository due to data protection policies of the study. Study data can only be accessed via the ODAP, a protected research environment. All data used in this study are available within ODAP and accessible under reasonable request. Data access criteria and information about how to request access is available online at ref. 76 . If criteria are met and a request is made, access can be gained by signing the eDRIS user agreement.
Code availability
Code was written within the ODAP, using R v4.2.0 and publicly available packages (‘data.table v1.14.2’, ‘EnvStats v2.7.0’, ‘tidyverse v1.3.2’, ‘lme4 v1.1-32’, ‘caret v6.0-93’, ‘glmnet v4.1-6’, ‘mdatools v0.14.0’, ‘ggpubbr v0.4.0’, ‘ggplot2 v3.3.6’, ‘bootnet v1.5.6’ and ‘qgraph v1.9.8’ packages). No new algorithms or functions were created and code used in-built functions in listed packages available on CRAN. The code used to generate data and to analyze data is publicly available at https://github.com/isaric4c/wiki/wiki/ISARIC ; https://github.com/SurgicalInformatics/cocin_cc and https://github.com/ClaudiaEfstath/PHOSP_Olink_NatImm .
Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21 , 133–146 (2023).
Article CAS PubMed PubMed Central Google Scholar
Antonelli, M., Pujol, J. C., Spector, T. D., Ourselin, S. & Steves, C. J. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet 399 , 2263–2264 (2022).
Talla, A. et al. Persistent serum protein signatures define an inflammatory subcategory of long COVID. Nat. Commun. 14 , 3417 (2023).
Captur, G. et al. Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection. EBioMedicine 85 , 104293 (2022).
Scott, N. A. et al. Monocyte migration profiles define disease severity in acute COVID-19 and unique features of long COVID. Eur. Respir. J. https://doi.org/10.1183/13993003.02226-2022 (2023).
Klein, J. et al. Distinguishing features of Long COVID identified through immune profiling. Nature https://doi.org/10.1038/s41586-023-06651-y (2023).
Article PubMed PubMed Central Google Scholar
Evans, R. A. et al. Clinical characteristics with inflammation profiling of long COVID and association with 1-year recovery following hospitalisation in the UK: a prospective observational study. Lancet Respir. Med . 10 , 761–775 (2022).
Article CAS Google Scholar
Houchen-Wolloff, L. et al. Joint patient and clinician priority setting to identify 10 key research questions regarding the long-term sequelae of COVID-19. Thorax 77 , 717–720 (2022).
Article PubMed Google Scholar
Marshall, J. C. et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 20 , e192–e197 (2020).
Post COVID-19 condition (long COVID). World Health Organization https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition#:~:text=Definition,months%20with%20no%20other%20explanation (2022).
Peters, V. A., Joesting, J. J. & Freund, G. G. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immun. 32 , 1–8 (2013).
Article CAS PubMed Google Scholar
Luo, Z. et al. Monocytes augment inflammatory responses in human aortic valve interstitial cells via β2-integrin/ICAM-1-mediated signaling. Inflamm. Res. 71 , 681–694 (2022).
Bendall, L. J. & Bradstock, K. F. G-CSF: from granulopoietic stimulant to bone marrow stem cell mobilizing agent. Cytokine Growth Factor Rev. 25 , 355–367 (2014).
Ma, Y. J. et al. Soluble collectin-12 (CL-12) is a pattern recognition molecule initiating complement activation via the alternative pathway. J. Immunol. 195 , 3365–3373 (2015).
Laursen, N. S. et al. Functional and structural characterization of a potent C1q inhibitor targeting the classical pathway of the complement system. Front. Immunol. 11 , 1504 (2020).
Dejanovic, B. et al. Complement C1q-dependent excitatory and inhibitory synapse elimination by astrocytes and microglia in Alzheimer’s disease mouse models. Nat. Aging 2 , 837–850 (2022).
Xue, G., Hua, L., Zhou, N. & Li, J. Characteristics of immune cell infiltration and associated diagnostic biomarkers in ulcerative colitis: results from bioinformatics analysis. Bioengineered 12 , 252–265 (2021).
He, T. et al. Integrative computational approach identifies immune‐relevant biomarkers in ulcerative colitis. FEBS Open Bio. 12 , 500–515 (2022).
Sundin, J. et al. Fecal chromogranins and secretogranins are linked to the fecal and mucosal intestinal bacterial composition of IBS patients and healthy subjects. Sci. Rep. 8 , 16821 (2018).
Kriebel, M., Wuchter, J., Trinks, S. & Volkmer, H. Neurofascin: a switch between neuronal plasticity and stability. Int. J. Biochem. Cell Biol. 44 , 694–697 (2012).
Woo, W.-M. et al. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development 135 , 2747–2756 (2008).
Yadati, T., Houben, T., Bitorina, A. & Shiri-Sverdlov, R. The ins and outs of cathepsins: physiological function and role in disease management. Cells 9 , 1679 (2020).
Taquet, M. et al. Acute blood biomarker profiles predict cognitive deficits 6 and 12 months after COVID-19 hospitalization. Nat. Med. https://doi.org/10.1038/s41591-023-02525-y (2023).
Siggins, M. K. et al. Alternative pathway dysregulation in tissues drives sustained complement activation and predicts outcome across the disease course in COVID‐19. Immunology 168 , 473–492 (2023).
Pol, J. G., Caudana, P., Paillet, J., Piaggio, E. & Kroemer, G. Effects of interleukin-2 in immunostimulation and immunosuppression. J. Exp. Med. 217 , e20191247 (2020).
Zhang, Y., Liu, Q., Yang, S. & Liao, Q. CD58 immunobiology at a glance. Front. Immunol. 12 , 705260 (2021).
Demydchuk, M. et al. Insights into Hunter syndrome from the structure of iduronate-2-sulfatase. Nat. Commun. 8 , 15786 (2017).
Wang, Z. et al. DNER promotes epithelial–mesenchymal transition and prevents chemosensitivity through the Wnt/β-catenin pathway in breast cancer. Cell Death Dis. 11 , 642 (2020).
Bonilla, H. et al. Therapeutic trials for long COVID-19: a call to action from the interventions taskforce of the RECOVER initiative. Front. Immunol. 14 , 1129459 (2023).
Wik, L. et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol. Cell. Proteomics 20 , 100168 (2021).
Measuring protein biomarkers with Olink—technical comparisons and orthogonal validation. Olink Proteomics https://www.olink.com/content/uploads/2021/09/olink-technical-comparisons-and-orthogonal-validation-1118-v2.0.pdf (2021).
COVID-19 rapid guideline: managing the long-term effects of COVID-19. National Institute for Health and Care Excellence (NICE), Scottish Intercollegiate Guidelines Network (SIGN) and Royal College of General Practitioners (RCGP) https://www.nice.org.uk/guidance/ng188/resources/covid19-rapid-guideline-managing-the-longterm-effects-of-covid19-pdf-51035515742 (2022).
Long COVID or post-COVID conditions. Centers for Disease Control and Prevention https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html#:~:text=Long%20COVID%20is%20broadly%20defined,after%20acute%20COVID%2D19%20infection (2023).
Firinguetti, L., Kibria, G. & Araya, R. Study of partial least squares and ridge regression methods. Commun. Stat. Simul. Comput 46 , 6631–6644 (2017).
Article Google Scholar
Mortensen, L. J., Kreiner-Moller, E., Hakonarson, H., Bonnelykke, K. & Bisgaard, H. The PCDH1 gene and asthma in early childhood. Eur. Respir. J. 43 , 792–800 (2014).
Tong, Y. et al. The RNFT2/IL-3Rα axis regulates IL-3 signaling and innate immunity. JCI Insight 5 , e133652 (2020).
Wu, Y. et al. Effect of ISM1 on the immune microenvironment and epithelial-mesenchymal transition in colorectal cancer. Front. Cell Dev. Biol. 9 , 681240 (2021).
Luo, G. G. & Ou, J. J. Oncogenic viruses and cancer. Virol. Sin. 30 , 83–84 (2015).
Klein, S. L. & Flanagan, K. L. Sex differences in immune responses. Nat. Rev. Immunol. 16 , 626–638 (2016).
Gaebler, C. et al. Evolution of antibody immunity to SARS-CoV-2. Nature 591 , 639–644 (2021).
Bussani, R. et al. Persistent SARS‐CoV‐2 infection in patients seemingly recovered from COVID‐19. J. Pathol. 259 , 254–263 (2023).
Woodruff, M. C. et al. Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat. Commun. 14 , 4201 (2023).
Lammi, V. et al. Genome-wide association study of long COVID. Preprint at medRxiv https://doi.org/10.1101/2023.06.29.23292056 (2023).
Ismailova, A. et al. Identification of a forkhead box protein transcriptional network induced in human neutrophils in response to inflammatory stimuli. Front. Immunol. 14 , 1123344 (2023).
Beurskens, F. J., van Schaarenburg, R. A. & Trouw, L. A. C1q, antibodies and anti-C1q autoantibodies. Mol. Immunol. 68 , 6–13 (2015).
Cervia-Hasler, C. et al. Persistent complement dysregulation with signs of thromboinflammation in active long Covid. Science 383 , eadg7942 (2024).
Morgan, B. P. & Harris, C. L. Complement, a target for therapy in inflammatory and degenerative diseases. Nat. Rev. Drug Discov. 14 , 857–877 (2015).
Toshner, M. R. et al. Apixaban following discharge in hospitalised adults with COVID-19: preliminary results from a multicentre, open-label, randomised controlled platform clinical trial. Preprint at medRxiv , https://doi.org/10.1101/2022.12.07.22283175 (2022).
Su, Y. et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185 , 881–895.e20 (2022).
Branchfield, K. et al. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351 , 707–710 (2016).
Rivera-Torruco, G. et al. Isthmin 1 identifies a subset of lung hematopoietic stem cells and it is associated with systemic inflammation. J. Immunol. 202 , 118.18 (2019).
Swank, Z. et al. Persistent circulating severe acute respiratory syndrome coronavirus 2 spike is associated with post-acute coronavirus disease 2019 sequelae. Clin. Infect. Dis. 76 , e487–e490 (2023).
Peluso, M. J. et al. Chronic viral coinfections differentially affect the likelihood of developing long COVID. J. Clin. Invest. 133 , e163669 (2023).
Bellocchi, C. et al. The interplay between autonomic nervous system and inflammation across systemic autoimmune diseases. Int. J. Mol. Sci. 23 , 2449 (2022).
Raman, B. et al. Multiorgan MRI findings after hospitalisation with COVID-19 in the UK (C-MORE): a prospective, multicentre, observational cohort study. Lancet Respir. Med 11 , 1003–1019 (2023).
Fernández-Castañeda, A. et al. Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell 185 , 2452–2468.e16 (2022).
Dantzer, R., O’Connor, J. C., Freund, G. G., Johnson, R. W. & Kelley, K. W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9 , 46–56 (2008).
Broughton, S. E. et al. Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody. Cell Rep. 8 , 410–419 (2014).
Ley, K., Miller, Y. I. & Hedrick, C. C. Monocyte and macrophage dynamics during atherogenesis. Arterioscler. Thromb. Vasc. Biol. 31 , 1506–1516 (2011).
Iosef, C. et al. Plasma proteome of long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function. J. Transl. Med. 21 , 377 (2023).
Santopaolo, M. et al. Prolonged T-cell activation and long COVID symptoms independently associate with severe COVID-19 at 3 months. eLife 12 , e85009 (2023).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 , 1–22 (2010).
Voiriot, G. et al. Chronic critical illness and post-intensive care syndrome: from pathophysiology to clinical challenges. Ann. Intensive Care 12 , 58 (2022).
Evans, R. A. et al. Physical, cognitive, and mental health impacts of COVID-19 after hospitalisation (PHOSP-COVID): a UK multicentre, prospective cohort study. Lancet Respir. Med 9 , 1275–1287 (2021).
Docherty, A. B. et al. Features of 20,133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ https://doi.org/10.1136/bmj.m1985 (2020).
Elneima, O. et al. Cohort profile: post-hospitalisation COVID-19 study (PHOSP-COVID). Preprint at medRxiv https://doi.org/10.1101/2023.05.08.23289442 (2023).
Liew, F. et al. SARS-CoV-2-specific nasal IgA wanes 9 months after hospitalisation with COVID-19 and is not induced by subsequent vaccination. EBioMedicine 87 , 104402 (2023).
Ascough, S. et al. Divergent age-related humoral correlates of protection against respiratory syncytial virus infection in older and young adults: a pilot, controlled, human infection challenge model. Lancet Healthy Longev. 3 , e405–e416 (2022).
Guvenel, A. et al. Epitope-specific airway-resident CD4+ T cell dynamics during experimental human RSV infection. J. Clin. Invest. 130 , 523–538 (2019).
Article PubMed Central Google Scholar
Greenwood, C. J. et al. A comparison of penalised regression methods for informing the selection of predictive markers. PLoS ONE 15 , e0242730 (2020).
Higham, A. et al. Leukotriene B4 levels in sputum from asthma patients. ERJ Open Res. 2 , 00088–02015 (2016).
SARS-CoV-2 spike kit. MSD https://www.mesoscale.com/~/media/files/product%20inserts/s-plex%20sars-cov-2%20spike%20kit%20product%20insert.pdf (2023).
Ren, A. et al. Ultrasensitive assay for saliva-based SARS-CoV-2 antigen detection. Clin. Chem. Lab. Med. 60 , 771–777 (2022).
Breheny, P. & Huang, J. Penalized methods for bi-level variable selection. Stat. Interface 2 , 369–380 (2009).
Thwaites, R. S. et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 6 , eabg9873 (2021).
Resources. PHOSP-COVID https://phosp.org/resource/ (2022).
Download references
Acknowledgements
This research used data assets made available by ODAP as part of the Data and Connectivity National Core Study, led by Health Data Research UK in partnership with the Office for National Statistics and funded by UK Research and Innovation (grant ref. MC_PC_20058). This work is supported by the following grants: the PHOSP-COVD study is jointly funded by UK Research and Innovation and National Institute of Health and Care Research (NIHR; grant references MR/V027859/1 and COV0319). ISARIC4C is supported by grants from the National Institute for Health and Care Research (award CO-CIN-01) and the MRC (grant MC_PC_19059) Liverpool Experimental Cancer Medicine Centre provided infrastructure support for this research (grant reference C18616/A25153). Other grants that have supported this work include the UK Coronavirus Immunology Consortium (funder reference 1257927), the Imperial Biomedical Research Centre (NIHR Imperial BRC, grant IS-BRC-1215-20013), the Health Protection Research Unit in Respiratory Infections at Imperial College London and NIHR Health Protection Research Unit in Emerging and Zoonotic Infections at University of Liverpool, both in partnership with Public Health England, (NIHR award 200907), Wellcome Trust and Department for International Development (215091/Z/18/Z), Health Data Research UK (grant code 2021.0155), MRC (grant code MC_UU_12014/12) and NIHR Clinical Research Network for providing infrastructure support for this research. We also acknowledge the support of the MRC EMINENT Network (MR/R502121/1), which is cofunded by GSK, the Comprehensive Local Research Networks, the MRC HIC-Vac network (MR/R005982/1) and the RSV Consortium in Europe Horizon 2020 Framework Grant 116019. F.L. is supported by an MRC clinical training fellowship (award MR/W000970/1). C.E. is funded by NIHR (grant P91258-4). L.-P.H. is supported by Oxford NIHR Biomedical Research Centre. A.A.R.T. is supported by a British Heart Foundation (BHF) Intermediate Clinical Fellowship (FS/18/13/33281). S.L.R.-J. receives support from UK Research and Innovation (UKRI), Global Challenges Research Fund (GCRF), Rosetrees Trust, British HIV association (BHIVA), European & Developing Countries Clinical Trials Partnership (EDCTP) and Globvac. J.D.C. has grants from AstraZeneca, Boehringer Ingelheim, GSK, Gilead Sciences, Grifols, Novartis and Insmed. R.A.E. holds a NIHR Clinician Scientist Fellowship (CS-2016-16-020). A. Horsley is currently supported by UK Research and Innovation, NIHR and NIHR Manchester BRC. B.R. receives support from BHF Oxford Centre of Research Excellence, NIHR Oxford BRC and MRC. D.G.W. is supported by an NIHR Advanced Fellowship. A. Ho has received support from MRC and for the Coronavirus Immunology Consortium (MR/V028448/1). L.T. is supported by the US Food and Drug Administration Medical Countermeasures Initiative contract 75F40120C00085 and the National Institute for Health Research Health Protection Research Unit in Emerging and Zoonotic Infections (NIHR200907) at the University of Liverpool in partnership with UK Health Security Agency (UK-HSA), in collaboration with Liverpool School of Tropical Medicine and the University of Oxford. L.V.W. has received support from UKRI, GSK/Asthma and Lung UK and NIHR for this study. M.G.S. has received support from NIHR UK, MRC UK and Health Protection Research Unit in Emerging and Zoonotic Infections, University of Liverpool. J.K.B. is supported by the Wellcome Trust (223164/Z/21/Z) and UKRI (MC_PC_20004, MC_PC_19025, MC_PC_1905, MRNO2995X/1 and MC_PC_20029). The funders were not involved in the study design, interpretation of data or writing of this manuscript. The views expressed are those of the authors and not necessarily those of the Department of Health and Social Care (DHSC), the Department for International Development (DID), NIHR, MRC, the Wellcome Trust, UK-HSA, the National Health Service or the Department of Health. P.J.M.O. is supported by a NIHR Senior Investigator Award (award 201385). We thank all the participants and their families. We thank the many research administrators, health-care and social-care professionals who contributed to setting up and delivering the PHOSP-COVID study at all of the 65 NHS trusts/health boards and 25 research institutions across the United Kingdom, as well as those who contributed to setting up and delivering the ISARIC4C study at 305 NHS trusts/health boards. We also thank all the supporting staff at the NIHR Clinical Research Network, Health Research Authority, Research Ethics Committee, Department of Health and Social Care, Public Health Scotland and Public Health England. We thank K. Holmes at the NIHR Office for Clinical Research Infrastructure for her support in coordinating the charities group. The PHOSP-COVID industry framework was formed to provide advice and support in commercial discussions, and we thank the Association of the British Pharmaceutical Industry as well the NIHR Office for Clinical Research Infrastructure for coordinating this. We are very grateful to all the charities that have provided insight to the study: Action Pulmonary Fibrosis, Alzheimer’s Research UK, Asthma and Lung UK, British Heart Foundation, Diabetes UK, Cystic Fibrosis Trust, Kidney Research UK, MQ Mental Health, Muscular Dystrophy UK, Stroke Association Blood Cancer UK, McPin Foundations and Versus Arthritis. We thank the NIHR Leicester Biomedical Research Centre patient and public involvement group and Long Covid Support. We also thank G. Khandaker and D. C. Newcomb who provided valuable feedback on this work. Extended Data Fig. 10 was created using Biorender.
Author information
These authors contributed equally: Felicity Liew, Claudia Efstathiou, Ryan S. Thwaites, Peter J. M. Openshaw.
Authors and Affiliations
National Heart and Lung Institute, Imperial College London, London, UK
Felicity Liew, Claudia Efstathiou, Sara Fontanella, Dawid Swieboda, Jasmin K. Sidhu, Stephanie Ascough, Onn Min Kon, Luke S. Howard, Jennifer K. Quint, Christopher Chiu, Ryan S. Thwaites, Peter J. M. Openshaw, Jake Dunning & Peter J. M. Openshaw
Institute for Lung Health, Leicester NIHR Biomedical Research Centre, University of Leicester, Leicester, UK
Matthew Richardson, Ruth Saunders, Olivia C. Leavy, Omer Elneima, Hamish J. C. McAuley, Amisha Singapuri, Marco Sereno, Victoria C. Harris, Neil J. Greening, Rachael A. Evans, Louise V. Wain, Christopher Brightling & Ananga Singapuri
NIHR Health Protection Research Unit in Emerging and Zoonotic Infections, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK
Shona C. Moore, Daniel G. Wootton, Malcolm G. Semple, Lance Turtle, William A. Paxton & Georgios Pollakis
The Imperial Clinical Respiratory Research Unit, Imperial College NHS Trust, London, UK
Noura Mohamed
Cardiovascular Research Team, Imperial College Healthcare NHS Trust, London, UK
Jose Nunag & Clara King
Department of Population Health Sciences, University of Leicester, Leicester, UK
Olivia C. Leavy, Louise V. Wain & Beatriz Guillen-Guio
NIHR Leicester Biomedical Research Centre, University of Leicester, Leicester, UK
Aarti Shikotra
Centre for Exercise and Rehabilitation Science, NIHR Leicester Biomedical Research Centre-Respiratory, University of Leicester, Leicester, UK
Linzy Houchen-Wolloff
Usher Institute, University of Edinburgh, Edinburgh, UK
Nazir I. Lone, Luke Daines, Annemarie B. Docherty, Nazir I. Lone, Matthew Thorpe, Annemarie B. Docherty, Thomas M. Drake, Cameron J. Fairfield, Ewen M. Harrison, Stephen R. Knight, Kenneth A. Mclean, Derek Murphy, Lisa Norman, Riinu Pius & Catherine A. Shaw
Centre for Medical Informatics, The Usher Institute, University of Edinburgh, Edinburgh, UK
Matthew Thorpe, Annemarie B. Docherty, Ewen M. Harrison, J. Kenneth Baillie, Sarah L. Rowland-Jones, A. A. Roger Thompson & Thushan de Silva
Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, UK
A. A. Roger Thompson, Sarah L. Rowland-Jones, Thushan I. de Silva & James D. Chalmers
University of Dundee, Ninewells Hospital and Medical School, Dundee, UK
James D. Chalmers & Ling-Pei Ho
MRC Human Immunology Unit, University of Oxford, Oxford, UK
Ling-Pei Ho & Alexander Horsley
Division of Infection, Immunity and Respiratory Medicine, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK
Alexander Horsley & Betty Raman
Radcliffe Department of Medicine, University of Oxford, Oxford, UK
Betty Raman & Krisnah Poinasamy
Asthma + Lung UK, London, UK
Krisnah Poinasamy & Michael Marks
Department of Clinical Research, London School of Hygiene and Tropical Medicine, London, UK
Michael Marks
Hospital for Tropical Diseases, University College London Hospital, London, UK
Division of Infection and Immunity, University College London, London, UK
Michael Marks & Mahdad Noursadeghi
MRC Centre for Virus Research, School of Infection and Immunity, University of Glasgow, Glasgow, UK
Antonia Ho & William Greenhalf
Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, UK
William Greenhalf & J. Kenneth Baillie
The Roslin Institute, University of Edinburgh, Edinburgh, UK
J. Kenneth Baillie, J. Kenneth Baillie, Sara Clohisey, Fiona Griffiths, Ross Hendry, Andrew Law & Wilna Oosthuyzen
Pandemic Science Hub, University of Edinburgh, Edinburgh, UK
J. Kenneth Baillie
The Pandemic Institute, University of Liverpool, Liverpool, UK
Malcolm G. Semple & Lance Turtle
University of Manchester, Manchester, UK
Kathryn Abel, Perdita Barran, H. Chinoy, Bill Deakin, M. Harvie, C. A. Miller, Stefan Stanel & Drupad Trivedi
Intensive Care Unit, Royal Infirmary of Edinburgh, Edinburgh, UK
Kathryn Abel & J. Kenneth Baillie
North Bristol NHS Trust and University of Bristol, Bristol, UK
H. Adamali, David Arnold, Shaney Barratt, A. Dipper, Sarah Dunn, Nick Maskell, Anna Morley, Leigh Morrison, Louise Stadon, Samuel Waterson & H. Welch
University of Edinburgh, Manchester, UK
Davies Adeloye, D. E. Newby, Riinu Pius, Igor Rudan, Manu Shankar-Hari, Catherine Sudlow, Sarah Walmsley & Bang Zheng
King’s College Hospital NHS Foundation Trust and King’s College London, London, UK
Oluwaseun Adeyemi, Rita Adrego, Hosanna Assefa-Kebede, Jonathon Breeze, S. Byrne, Pearl Dulawan, Amy Hoare, Caroline Jolley, Abigail Knighton, M. Malim, Sheetal Patale, Ida Peralta, Natassia Powell, Albert Ramos, K. Shevket, Fabio Speranza & Amelie Te
Guy’s and St Thomas’ NHS Foundation Trust, London, UK
Laura Aguilar Jimenez, Gill Arbane, Sarah Betts, Karen Bisnauthsing, A. Dewar, Nicholas Hart, G. Kaltsakas, Helen Kerslake, Murphy Magtoto, Philip Marino, L. M. Martinez, Marlies Ostermann, Jennifer Rossdale & Teresa Solano
Royal Free London NHS Foundation Trust, London, UK
Shanaz Ahmad, Simon Brill, John Hurst, Hannah Jarvis, C. Laing, Lai Lim, S. Mandal, Darwin Matila, Olaoluwa Olaosebikan & Claire Singh
University Hospital Birmingham NHS Foundation Trust and University of Birmingham, Birmingham, UK
N. Ahmad Haider, Catherine Atkin, Rhiannon Baggott, Michelle Bates, A. Botkai, Anna Casey, B. Cooper, Joanne Dasgin, Camilla Dawson, Katharine Draxlbauer, N. Gautam, J. Hazeldine, T. Hiwot, Sophie Holden, Karen Isaacs, T. Jackson, Vicky Kamwa, D. Lewis, Janet Lord, S. Madathil, C. McGee, K. Mcgee, Aoife Neal, Alex Newton-Cox, Joseph Nyaboko, Dhruv Parekh, Z. Peterkin, H. Qureshi, Liz Ratcliffe, Elizabeth Sapey, J. Short, Tracy Soulsby, J. Stockley, Zehra Suleiman, Tamika Thompson, Maximina Ventura, Sinead Walder, Carly Welch, Daisy Wilson, S. Yasmin & Kay Por Yip
Stroke Association, London, UK
Rubina Ahmed & Richard Francis
University College London Hospital and University College London, London, UK
Nyarko Ahwireng, Dongchun Bang, Donna Basire, Jeremy Brown, Rachel Chambers, A. Checkley, R. Evans, M. Heightman, T. Hillman, Joseph Jacob, Roman Jastrub, M. Lipman, S. Logan, D. Lomas, Marta Merida Morillas, Hannah Plant, Joanna Porter, K. Roy & E. Wall
Oxford University Hospitals NHS Foundation Trust and University of Oxford, Oxford, UK
Mark Ainsworth, Asma Alamoudi, Angela Bloss, Penny Carter, M. Cassar, Jin Chen, Florence Conneh, T. Dong, Ranuromanana Evans, V. Ferreira, Emily Fraser, John Geddes, F. Gleeson, Paul Harrison, May Havinden-Williams, P. Jezzard, Ivan Koychev, Prathiba Kurupati, H. McShane, Clare Megson, Stefan Neubauer, Debby Nicoll, C. Nikolaidou, G. Ogg, Edmund Pacpaco, M. Pavlides, Yanchun Peng, Nayia Petousi, John Pimm, Najib Rahman, M. J. Rowland, Kathryn Saunders, Michael Sharpe, Nick Talbot, E. M. Tunnicliffe & C. Xie
St George’s University Hospitals NHS Foundation Trust, London, UK
Mariam Ali, Raminder Aul, A. Dunleavy, D. Forton, Mark Mencias, N. Msimanga, T. Samakomva, Sulman Siddique, Vera Tavoukjian & J. Teixeira
University Hospitals of Leicester NHS Trust and University of Leicester, Leicester, UK
M. Aljaroof, Natalie Armstrong, H. Arnold, Hnin Aung, Majda Bakali, M. Bakau, E. Baldry, Molly Baldwin, Charlotte Bourne, Michelle Bourne, Nigel Brunskill, P. Cairns, Liesel Carr, Amanda Charalambou, C. Christie, Melanie Davies, Enya Daynes, Sarah Diver, Rachael Dowling, Sarah Edwards, C. Edwardson, H. Evans, J. Finch, Sarah Glover, Nicola Goodman, Bibek Gooptu, Kate Hadley, Pranab Haldar, Beverley Hargadon, W. Ibrahim, L. Ingram, Kamlesh Khunti, A. Lea, D. Lee, Gerry McCann, P. McCourt, Teresa Mcnally, George Mills, Will Monteiro, Manish Pareek, S. Parker, Anne Prickett, I. N. Qureshi, A. Rowland, Richard Russell, Salman Siddiqui, Sally Singh, J. Skeemer, M. Soares, E. Stringer, T. Thornton, Martin Tobin, T. J. C. Ward, F. Woodhead, Tom Yates & A. J. Yousuf
University of Exeter, Exeter, UK
Louise Allan, Clive Ballard & Andrew McGovern
University of Leicester, Leicester, UK
Richard Allen, Michelle Bingham, Terry Brugha, Selina Finney, Rob Free, Don Jones, Claire Lawson, Daniel Lozano-Rojas, Gardiner Lucy, Alistair Moss, Elizabeta Mukaetova-Ladinska, Petr Novotny, Kimon Ntotsis, Charlotte Overton, John Pearl, Tatiana Plekhanova, M. Richardson, Nilesh Samani, Jack Sargant, Ruth Saunders, M. Sharma, Mike Steiner, Chris Taylor, Sarah Terry, C. Tong, E. Turner, J. Wormleighton & Bang Zhao
Liverpool University Hospitals NHS Foundation Trust and University of Liverpool, Liverpool, UK
Lisa Allerton, Ann Marie Allt, M. Beadsworth, Anthony Berridge, Jo Brown, Shirley Cooper, Andy Cross, Sylviane Defres, S. L. Dobson, Joanne Earley, N. French, Kera Hainey, Hayley Hardwick, Jenny Hawkes, Victoria Highett, Sabina Kaprowska, Angela Key, Lara Lavelle-Langham, N. Lewis-Burke, Gladys Madzamba, Flora Malein, Sophie Marsh, Chloe Mears, Lucy Melling, Matthew Noonan, L. Poll, James Pratt, Emma Richardson, Anna Rowe, Victoria Shaw, K. A. Tripp, Lilian Wajero, S. A. Williams-Howard, Dan Wootton & J. Wyles
Sherwood Forest Hospitals NHS Foundation Trust, Nottingham, UK
Lynne Allsop, Kaytie Bennett, Phil Buckley, Margaret Flynn, Mandy Gill, Camelia Goodwin, M. Greatorex, Heidi Gregory, Cheryl Heeley, Leah Holloway, Megan Holmes, John Hutchinson, Jill Kirk, Wayne Lovegrove, Terri Ann Sewell, Sarah Shelton, D. Sissons, Katie Slack, Susan Smith, D. Sowter, Sarah Turner, V. Whitworth & Inez Wynter
Nottingham University Hospitals NHS Trust and University of Nottingham, London, UK
Paula Almeida, Akram Hosseini, Robert Needham & Karen Shaw
Manchester University NHS Foundation Trust and University of Manchester, London, UK
Bashar Al-Sheklly, Cristina Avram, John Blaikely, M. Buch, N. Choudhury, David Faluyi, T. Felton, T. Gorsuch, Neil Hanley, Tracy Hussell, Zunaira Kausar, Natasha Odell, Rebecca Osbourne, Karen Piper Hanley, K. Radhakrishnan & Sue Stockdale
Imperial College London, London, UK
Danny Altmann, Anew Frankel, Luke S. Howard, Desmond Johnston, Liz Lightstone, Anne Lingford-Hughes, William Man, Steve McAdoo, Jane Mitchell, Philip Molyneaux, Christos Nicolaou, D. P. O’Regan, L. Price, Jennifer K. Quint, David Smith, Jonathon Valabhji, Simon Walsh, Martin Wilkins & Michelle Willicombe
Hampshire Hospitals NHS Foundation Trust, Basingstoke, UK
Maria Alvarez Corral, Ava Maria Arias, Emily Bevan, Denise Griffin, Jane Martin, J. Owen, Sheila Payne, A. Prabhu, Annabel Reed, Will Storrar, Nick Williams & Caroline Wrey Brown
British Heart Foundation, Birmingham, UK
Shannon Amoils
NHS Greater Glasgow and Clyde Health Board and University of Glasgow, Glasgow, UK
David Anderson, Neil Basu, Hannah Bayes, Colin Berry, Ammani Brown, Andrew Dougherty, K. Fallon, L. Gilmour, D. Grieve, K. Mangion, I. B. McInnes, A. Morrow, Kathryn Scott & R. Sykes
University of Oxford, Oxford, UK
Charalambos Antoniades, A. Bates, M. Beggs, Kamaldeep Bhui, Katie Breeze, K. M. Channon, David Clark, X. Fu, Masud Husain, Lucy Kingham, Paul Klenerman, Hanan Lamlum, X. Li, E. Lukaschuk, Celeste McCracken, K. McGlynn, R. Menke, K. Motohashi, T. E. Nichols, Godwin Ogbole, S. Piechnik, I. Propescu, J. Propescu, A. A. Samat, Z. B. Sanders, Louise Sigfrid & M. Webster
Belfast Health and Social Care Trust and Queen’s University Belfast, Belfast, UK
Cherie Armour, Vanessa Brown, John Busby, Bronwen Connolly, Thelma Craig, Stephen Drain, Liam Heaney, Bernie King, Nick Magee, E. Major, Danny McAulay, Lorcan McGarvey, Jade McGinness, Tunde Peto & Roisin Stone
Airedale NHS Foundation Trust, Keighley, UK
Lisa Armstrong, Brigid Hairsine, Helen Henson, Claire Kurasz, Alison Shaw & Liz Shenton
Wrightington Wigan and Leigh NHS Trust, Wigan, UK
A. Ashish, Josh Cooper & Emma Robinson
Leeds Teaching Hospitals and University of Leeds, Leeds, UK
Andrew Ashworth, Paul Beirne, Jude Clarke, C. Coupland, Matthhew Dalton, Clair Favager, Jodie Glossop, John Greenwood, Lucy Hall, Tim Hardy, Amy Humphries, Jennifer Murira, Dan Peckham, S. Plein, Jade Rangeley, Gwen Saalmink, Ai Lyn Tan, Elaine Wade, Beverley Whittam, Nicola Window & Janet Woods
University of Liverpool, Liverpool, UK
M. Ashworth, D. Cuthbertson, G. Kemp, Anne McArdle, Benedict Michael, Will Reynolds, Lisa Spencer, Ben Vinson, Katie A. Ahmed, Jane A. Armstrong, Milton Ashworth, Innocent G. Asiimwe, Siddharth Bakshi, Samantha L. Barlow, Laura Booth, Benjamin Brennan, Katie Bullock, Nicola Carlucci, Emily Cass, Benjamin W. A. Catterall, Jordan J. Clark, Emily A. Clarke, Sarah Cole, Louise Cooper, Helen Cox, Christopher Davis, Oslem Dincarslan, Alejandra Doce Carracedo, Chris Dunn, Philip Dyer, Angela Elliott, Anthony Evans, Lorna Finch, Lewis W. S. Fisher, Lisa Flaherty, Terry Foster, Isabel Garcia-Dorival, Philip Gunning, Catherine Hartley, Karl Holden, Anthony Holmes, Rebecca L. Jensen, Christopher B. Jones, Trevor R. Jones, Shadia Khandaker, Katharine King, Robyn T. Kiy, Chrysa Koukorava, Annette Lake, Suzannah Lant, Diane Latawiec, Lara Lavelle-Langham, Daniella Lefteri, Lauren Lett, Lucia A. Livoti, Maria Mancini, Hannah Massey, Nicole Maziere, Sarah McDonald, Laurence McEvoy, John McLauchlan, Soeren Metelmann, Nahida S. Miah, Joanna Middleton, Joyce Mitchell, Ellen G. Murphy, Rebekah Penrice-Randal, Jack Pilgrim, Tessa Prince, P. Matthew Ridley, Debby Sales, Rebecca K. Shears, Benjamin Small, Krishanthi S. Subramaniam, Agnieska Szemiel, Aislynn Taggart, Jolanta Tanianis-Hughes, Jordan Thomas, Erwan Trochu, Libby van Tonder, Eve Wilcock & J. Eunice Zhang
University College London, London, UK
Shahab Aslani, Amita Banerjee, R. Batterham, Gabrielle Baxter, Robert Bell, Anthony David, Emma Denneny, Alun Hughes, W. Lilaonitkul, P. Mehta, Ashkan Pakzad, Bojidar Rangelov, B. Williams, James Willoughby & Moucheng Xu
Hull University Teaching Hospitals NHS Trust and University of Hull, Hull, UK
Paul Atkin, K. Brindle, Michael Crooks, Katie Drury, Nicholas Easom, Rachel Flockton, L. Holdsworth, A. Richards, D. L. Sykes, Susannah Thackray-Nocera & C. Wright
East Kent Hospitals University NHS Foundation Trust, Canterbury, UK
Liam Austin, Eva Beranova, Tracey Cosier, Joanne Deery, Tracy Hazelton, Carly Price, Hazel Ramos, Reanne Solly, Sharon Turney & Heather Weston
Baillie Gifford Pandemic Science Hub, Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
Nikos Avramidis, J. Kenneth Baillie, Erola Pairo-Castineira & Konrad Rawlik
Roslin Institute, University of Edinburgh, Edinburgh, UK
Nikos Avramidis, J. Kenneth Baillie & Erola Pairo-Castineira
Newcastle upon Tyne Hospitals NHS Foundation Trust and University of Newcastle, Newcastle upon Tyne, UK
A. Ayoub, J. Brown, G. Burns, Gareth Davies, Anthony De Soyza, Carlos Echevarria, Helen Fisher, C. Francis, Alan Greenhalgh, Philip Hogarth, Joan Hughes, Kasim Jiwa, G. Jones, G. MacGowan, D. Price, Avan Sayer, John Simpson, H. Tedd, S. Thomas, Sophie West, M. Witham, S. Wright & A. Young
East Cheshire NHS Trust, Macclesfield, UK
Marta Babores, Maureen Holland, Natalie Keenan, Sharlene Shashaa & Helen Wassall
Sheffield Teaching NHS Foundation Trust and University of Sheffield, Sheffield, UK
J. Bagshaw, M. Begum, K. Birchall, Robyn Butcher, H. Carborn, Flora Chan, Kerry Chapman, Yutung Cheng, Luke Chetham, Cameron Clark, Zach Coburn, Joby Cole, Myles Dixon, Alexandra Fairman, J. Finnigan, H. Foot, David Foote, Amber Ford, Rebecca Gregory, Kate Harrington, L. Haslam, L. Hesselden, J. Hockridge, Ailsa Holbourn, B. Holroyd-Hind, L. Holt, Alice Howell, E. Hurditch, F. Ilyas, Claire Jarman, Allan Lawrie, Ju Hee Lee, Elvina Lee, Rebecca Lenagh, Alison Lye, Irene Macharia, M. Marshall, Angeline Mbuyisa, J. McNeill, Sharon Megson, J. Meiring, L. Milner, S. Misra, Helen Newell, Tom Newman, C. Norman, Lorenza Nwafor, Dibya Pattenadk, Megan Plowright, Julie Porter, Phillip Ravencroft, C. Roddis, J. Rodger, Peter Saunders, J. Sidebottom, Jacqui Smith, Laurie Smith, N. Steele, G. Stephens, R. Stimpson, B. Thamu, N. Tinker, Kim Turner, Helena Turton, Phillip Wade, S. Walker, James Watson, Imogen Wilson & Amira Zawia
University of Nottingham, Nottingham, UK
David Baguley, Chris Coleman, E. Cox, Laura Fabbri, Susan Francis, Ian Hall, E. Hufton, Simon Johnson, Fasih Khan, Paaig Kitterick, Richard Morriss, Nick Selby, Iain Stewart & Louise Wright
Wirral University Teaching Hospital, Wirral, UK
Elisabeth Bailey, Anne Reddington & Andrew Wight
MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Edinburgh, UK
University of Swansea, Swansea, UK
University of Southampton, London, UK
David Baldwin, P. C. Calder, Nathan Huneke & Gemma Simons
Royal Brompton and Harefield Clinical Group, Guy’s and St Thomas’ NHS Foundation Trust, London, UK
R. E. Barker, Daniele Cristiano, N. Dormand, P. George, Mahitha Gummadi, S. Kon, Kamal Liyanage, C. M. Nolan, B. Patel, Suhani Patel, Oliver Polgar, L. Price, P. Shah, Suver Singh & J. A. Walsh
York and Scarborough NHS Foundation Trust, York, UK
Laura Barman, Claire Brookes, K. Elliott, L. Griffiths, Zoe Guy, Kate Howard, Diana Ionita, Heidi Redfearn, Carol Sarginson & Alison Turnbull
NHS Highland, Inverness, UK
Fiona Barrett, A. Donaldson & Beth Sage
Royal Papworth Hospital NHS Foundation Trust, Cambridge, UK
Helen Baxendale, Lucie Garner, C. Johnson, J. Mackie, Alice Michael, J. Newman, Jamie Pack, K. Paques, H. Parfrey, J. Parmar & A. Reddy
University Hospitals of Derby and Burton, Derby, UK
Paul Beckett, Caroline Dickens & Uttam Nanda
NHS Lanarkshire, Hamilton, UK
Murdina Bell, Angela Brown, M. Brown, R. Hamil, Karen Leitch, L. Macliver, Manish Patel, Jackie Quigley, Andrew Smith & B. Welsh
Cambridge University Hospitals NHS Foundation Trust, NIHR Cambridge Clinical Research Facility and University of Cambridge, Cambridge, UK
Areti Bermperi, Isabel Cruz, K. Dempsey, Anne Elmer, Jonathon Fuld, H. Jones, Sherly Jose, Stefan Marciniak, M. Parkes, Carla Ribeiro, Jessica Taylor, Mark Toshner, L. Watson & J. Worsley
Loughborough University, Loughborough, UK
Lettie Bishop & David Stensel
Betsi Cadwallader University Health Board, Bangor, UK
Annette Bolger, Ffyon Davies, Ahmed Haggar, Joanne Lewis, Arwel Lloyd, R. Manley, Emma McIvor, Daniel Menzies, K. Roberts, W. Saxon, David Southern, Christian Subbe & Victoria Whitehead
Nottingham University Hospitals NHS Trust and University of Nottingham, Nottingham, UK
Charlotte Bolton, J. Bonnington, Melanie Chrystal, Catherine Dupont, Paul Greenhaff, Ayushman Gupta, W. Jang, S. Linford, Laura Matthews, Athanasios Nikolaidis, Sabrina Prosper & Andrew Thomas
King’s College London, London, UK
Kate Bramham, M. Brown, Khalida Ismail, Tim Nicholson, Carmen Pariante, Claire Sharpe, Simon Wessely & J. Whitney
Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK
Lucy Brear, Karen Regan, Dinesh Saralaya & Kim Storton
South London and Maudsley NHS Foundation Trust and King’s College London, London, UK
G. Breen & M. Hotopf
London School of Hygiene and Tropical Medicine, London, UK
Andrew Briggs
Whittington Health NHS Trust, London, UK
E. Bright, P. Crisp, Ruvini Dharmagunawardena & M. Stern
Cardiff and Vale University Health Board, Cardiff, UK
Lauren Broad, Teriann Evans, Matthew Haynes, L. Jones, Lucy Knibbs, Alison McQueen, Catherine Oliver, Kerry Paradowski, Ramsey Sabit & Jenny Williams
Yeovil District Hospital NHS Foundation Trust, Yeovil, UK
Andrew Broadley
University of Birmingham, Birmingham, UK
Mattew Broome, Paul McArdle, Paul Moss, David Thickett, Rachel Upthegrove, Dan Wilkinson, David Wraith & Erin L. Aldera
BHF Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK
Anda Bularga
University of Cambridge, Cambridge, UK
Ed Bullmore, Jonathon Heeney, Claudia Langenberg, William Schwaeble, Charlotte Summers & J. Weir McCall
NIHR Leicester Biomedical Research Centre–Respiratory Patient and Public Involvement Group, Leicester, UK
Jenny Bunker, Rhyan Gill & Rashmita Nathu
Imperial College Healthcare NHS Trust and Imperial College London, London, UK
L. Burden, Ellen Calvelo, Bethany Card, Caitlin Carr, Edwin Chilvers, Donna Copeland, P. Cullinan, Patrick Daly, Lynsey Evison, Tamanah Fayzan, Hussain Gordon, Sulaimaan Haq, Gisli Jenkins, Clara King, Onn Min Kon, Katherine March, Myril Mariveles, Laura McLeavey, Silvia Moriera, Unber Munawar, Uchechi Nwanguma, Lorna Orriss-Dib, Alexandra Ross, Maura Roy, Emily Russell, Katherine Samuel, J. Schronce, Neil Simpson, Lawrence Tarusan, David Thomas, Chloe Wood & Najira Yasmin
Harrogate and District NHD Foundation Trust, Harrogate, UK
Tracy Burdett, James Featherstone, Cathy Lawson, Alison Layton, Clare Mills & Lorraine Stephenson
Newcastle University/Chair of NIHR Dementia TRC, Newcastle, UK
Oxford University Hospitals NHS Foundation Trust, Oxford, UK
A. Burns & N. Kanellakis
Tameside and Glossop Integrated Care NHS Foundation Trust, Ashton-under-Lyne, UK
Al-Tahoor Butt, Martina Coulding, Heather Jones, Susan Kilroy, Jacqueline McCormick, Jerome McIntosh, Heather Savill, Victoria Turner & Joanne Vere
University of Oxford, Nuffield Department of Medicine, Oxford, UK
University of Glasgow, Glasgow, UK
Jonathon Cavanagh, S. MacDonald, Kate O’Donnell, John Petrie, Naveed Sattar & Mark Spears
United Lincolnshire Hospitals NHS Trust, Grantham, UK
Manish Chablani & Lynn Osborne
Department of Psychological Medicine, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK
Trudie Chalder
University Hospital of South Manchester NHS Foundation Trust, Manchester, UK
N. Chaudhuri
University Hospital Southampton NHS Foundation Trust and University of Southampton, Southampton, UK
Caroline Childs, R. Djukanovic, S. Fletcher, Matt Harvey, Mark Jones, Elizabeth Marouzet, B. Marshall, Reena Samuel, T. Sass, Tim Wallis & Helen Wheeler
King’s College Hospital/Guy’s and St Thomas’ NHS FT, London, UK
A. Chiribiri & C. O’Brien
Barts Health NHS Trust, London, UK
K. Chong-James, C. David, W. Y. James, Paul Pfeffer & O. Zongo
NHS Lothian and University of Edinburgh, Edinburgh, UK
Gaunab Choudhury, S. Clohisey, Andrew Deans, J. Furniss, Ewen Harrison, S. Kelly & Aziz Sheikh
School of Cardiovascular Medicine and Sciences. King’s College London, London, UK
Phillip Chowienczyk
Lewisham and Greenwich NHS Trust, London, UK
Hywel Dda University Health Board, Haverfordwest, UK
S. Coetzee, Kim Davies, Rachel Ann Hughes, Ronda Loosley, Heather McGuinness, Abdelrahman Mohamed, Linda O’Brien, Zohra Omar, Emma Perkins, Janet Phipps, Gavin Ross, Abigail Taylor, Helen Tench & Rebecca Wolf-Roberts
NHS Tayside and University of Dundee, Dundee, UK
David Connell, C. Deas, Anne Elliott, J. George, S. Mohammed, J. Rowland, A. R. Solstice, Debbie Sutherland & Caroline Tee
Swansea Bay University Health Board, Port Talbot, UK
Lynda Connor, Amanda Cook, Gwyneth Davies, Tabitha Rees, Favas Thaivalappil & Caradog Thomas
Faculty of Medicine, Nursing and Health Sciences, School of Biomedical Sciences, Monash University, Melbourne, Victoria, Australia
Eamon Coughlan
Rotherham NHS Foundation Trust, Rotherham, UK
Alison Daniels, Anil Hormis, Julie Ingham & Lisa Zeidan
Salford Royal NHS Foundation Trust, Salford, UK
P. Dark, Nawar Diar-Bakerly, D. Evans, E. Hardy, Alice Harvey, D. Holgate, Sean Knight, N. Mairs, N. Majeed, L. McMorrow, J. Oxton, Jessica Pendlebury, C. Summersgill, R. Ugwuoke & S. Whittaker
Cwm Taf Morgannwg University Health Board, Mountain Ash, UK
Ellie Davies, Cerys Evenden, Alyson Hancock, Kia Hancock, Ceri Lynch, Meryl Rees, Lisa Roche, Natalie Stroud & T. Thomas-Woods
Borders General Hospital, NHS Borders, Melrose, UK
Joy Dawson, Hosni El-Taweel & Leanne Robinson
Aneurin Bevan University Health Board, Caerleon, UK
Amanda Dell, Sara Fairbairn, Nancy Hawkings, Jill Haworth, Michaela Hoare, Victoria Lewis, Alice Lucey, Georgia Mallison, Heeah Nassa, Chris Pennington, Andrea Price, Claire Price, Andrew Storrie, Gemma Willis & Susan Young
University of Exeter Medical School, Exeter, UK
London North West University Healthcare NHS Trust, London, UK
Shalin Diwanji, Sambasivarao Gurram, Padmasayee Papineni, Sheena Quaid, Gerlynn Tiongson & Ekaterina Watson
Alzheimer’s Research UK, Cambridge, UK
Hannah Dobson
Health and Care Research Wales, Cardiff, UK
Yvette Ellis
University of Bristol, Bristol, UK
Jonathon Evans
University of Sheffield, Sheffield, UK
L. Finnigan, Laura Saunders & James Wild
Great Western Hospital Foundation Trust, Swindon, UK
Eva Fraile & Jacinta Ugoji
Royal Devon and Exeter NHS Trust, Barnstaple, UK
Michael Gibbons
Kettering General Hospital NHS Trust, Kettering, UK
Anne-Marie Guerdette, Melanie Hewitt, R. Reddy, Katie Warwick & Sonia White
NIHR Leicester Biomedical Research Centre, Leicester, UK
Beatriz Guillen-Guio
University of Leeds, Leeds, UK
Elspeth Guthrie & Max Henderson
Royal Surrey NHS Foundation Trust, Cranleigh, UK
Mark Halling-Brown & Katherine McCullough
Chesterfield Royal Hospital NHS Trust, Calow, UK
Edward Harris & Claire Sampson
Long Covid Support, London, UK
Claire Hastie, Natalie Rogers & Nikki Smith
King’s College Hospital, NHS Foundation Trust and King’s College London, London, UK
Department of Oncology and Metabolism, University of Sheffield, Sheffield, UK
Simon Heller
NIHR Office for Clinical Research Infrastructure, London, UK
Katie Holmes
Asthma UK and British Lung Foundation Partnership, London, UK
Ian Jarrold & Samantha Walker
North Middlesex University Hospital NHS Trust, London, UK
Bhagy Jayaraman & Tessa Light
Action for Pulmonary Fibrosis, Peterborough, UK
Cardiff University, National Centre for Mental Health, Cardiff, UK
McPin Foundation, London, UK
Thomas Kabir
Roslin Institute, The University of Edinburgh, Edinburgh, UK
Steven Kerr
The Hillingdon Hospitals NHS Foundation Trust, London, UK
Samantha Kon, G. Landers, Harpreet Lota, Mariam Nasseri & Sofiya Portukhay
Queen Mary University of London, London, UK
Ania Korszun
Swansea University, Swansea Welsh Network, Hywel Dda University Health Board, Swansea, UK
Royal Infirmary of Edinburgh, NHS Lothian, Edinburgh, UK
Nazir I. Lone
Barts Heart Centre, London, UK
Barts Health NHS Trust and Queen Mary University of London, London, UK
Adrian Martineau
Salisbury NHS Foundation Trust, Salisbury, UK
Wadzanai Matimba-Mupaya & Sophia Strong-Sheldrake
University of Newcastle, Newcastle, UK
Hamish McAllister-Williams, Stella-Maria Paddick, Anthony Rostron & John Paul Taylor
Gateshead NHS Trust, Gateshead, UK
W. McCormick, Lorraine Pearce, S. Pugmire, Wendy Stoker & Ann Wilson
Manchester Centre for Clinical Neurosciences, Salford Royal NHS Foundation Trust, Manchester, UK
Katherine McIvor
Kidney Research UK, Peterborough, UK
Aisling McMahon
NHS Dumfries and Galloway, Dumfries, UK
Michael McMahon & Paula Neill
Swansea University, Swansea, UK
MQ Mental Health Research, London, UK
Lea Milligan
BHF Centre for Cardiovascular Science, Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK
Nicholas Mills
Shropshire Community Health NHS Trust, Shropshire, UK
Sharon Painter, Johanne Tomlinson & Louise Warburton
Somerset NHS Foundation Trust, Taunton, UK
Sue Palmer, Dawn Redwood, Jo Tilley, Carinna Vickers & Tania Wainwright
Francis Crick Institute, London, UK
Markus Ralser
Manchester University NHD Foundation Trust, Manchester, UK
Pilar Rivera-Ortega
Diabetes UK, University of Glasgow, Glasgow, UK
Elizabeth Robertson
Barnsley Hospital NHS Foundation Trust, Barnsley, UK
Amy Sanderson
MRC–University of Glasgow Centre for Virus Research, Glasgow, UK
Janet Scott
Diabetes UK, London, UK
Kamini Shah
British Heart Foundation Centre, King’s College London, London, UK
King’s College Hospital NHS Foundation Trust, London, UK
University Hospitals Birmingham NHS Foundation Trust and University of Birmingham, Birmingham, UK
Institute of Cardiovascular and Medical Sciences, BHF Glasgow Cardiovascular Research Centre, University of Glasgow, Glasgow, UK
University College London NHS Foundation Trust, London and Barts Health NHS Trust, London, UK
Northumbria University, Newcastle upon Tyne, UK
Ioannis Vogiatzis
Swansea University and Swansea Welsh Network, Swansea, UK
N. Williams
DUK | NHS Digital, Salford Royal Foundation Trust, Salford, UK
Queen Alexandra Hospital, Portsmouth, UK
- Kayode Adeniji
Princess Royal Hospital, Haywards Heath, UK
Daniel Agranoff & Chi Eziefula
Bassetlaw Hospital, Bassetlaw, UK
Darent Valley Hospital, Dartford, UK
Queen Elizabeth the Queen Mother Hospital, Margate, UK
Ana Alegria
School of Informatics, University of Edinburgh, Edinburgh, UK
Beatrice Alex, Benjamin Bach & James Scott-Brown
North East and North Cumbria Ingerated, Newcastle upon Tyne, UK
Section of Biomolecular Medicine, Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Petros Andrikopoulos, Kanta Chechi, Marc-Emmanuel Dumas, Julian Griffin, Sonia Liggi & Zoltan Takats
Section of Genomic and Environmental Medicine, Respiratory Division, National Heart and Lung Institute, Imperial College London, London, UK
Petros Andrikopoulos, Marc-Emmanuel Dumas, Michael Olanipekun & Anthonia Osagie
John Radcliffe Hospital, Oxford, UK
Brian Angus
Royal Albert Edward Infirmary, Wigan, UK
Abdul Ashish
Manchester Royal Infirmary, Manchester, UK
Dougal Atkinson
MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, UK
Section of Molecular Virology, Imperial College London, London, UK
Wendy S. Barclay
Furness General Hospital, Barrow-in-Furness, UK
Shahedal Bari
Hull University Teaching Hospital Trust, Kingston upon Hull, UK
Gavin Barlow
Hillingdon Hospital, Hillingdon, UK
Stella Barnass
St Thomas’ Hospital, London, UK
Nicholas Barrett
Coventry and Warwickshire, Coventry, UK
Christopher Bassford
St Michael’s Hospital, Bristol, UK
Sneha Basude
Stepping Hill Hospital, Stockport, UK
David Baxter
Royal Liverpool University Hospital, Liverpool, UK
Michael Beadsworth
Bristol Royal Hospital Children’s, Bristol, UK
Jolanta Bernatoniene
Scarborough Hospital, Scarborough, UK
John Berridge
Golden Jubilee National Hospital, Clydebank, UK
Colin Berry
Liverpool Heart and Chest Hospital, Liverpool, UK
Nicola Best
Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
Debby Bogaert & Clark D. Russell
James Paget University Hospital, Great Yarmouth, UK
Pieter Bothma & Darell Tupper-Carey
Aberdeen Royal Infirmary, Aberdeen, UK
Robin Brittain-Long
Adamson Hospital, Cupar, UK
Naomi Bulteel
Royal Devon and Exeter Hospital, Exeter, UK
Worcestershire Royal Hospital, Worcester, UK
Andrew Burtenshaw
ISARIC Global Support Centre, Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Gail Carson, Laura Merson & Louise Sigfrid
Conquest Hospital, Hastings, UK
Vikki Caruth
The James Cook University Hospital, Middlesbrough, UK
David Chadwick
Dorset County Hospital, Dorchester, UK
Duncan Chambler
Antimicrobial Resistance and Hospital Acquired Infection Department, Public Health England, London, UK
Meera Chand
Department of Epidemiology and Biostatistics, School of Public Health, Faculty of Medicine, Imperial College London, London, UK
Kanta Chechi
Royal Bournemouth General Hospital, Bournemouth, UK
Harrogate Hospital, Harrogate, UK
Jenny Child
Royal Blackburn Teaching Hospital, Blackburn, UK
Srikanth Chukkambotla
Edinburgh Clinical Research Facility, University of Edinburgh, Edinburgh, UK
Richard Clark, Audrey Coutts, Lorna Donelly, Angie Fawkes, Tammy Gilchrist, Katarzyna Hafezi, Louise MacGillivray, Alan Maclean, Sarah McCafferty, Kirstie Morrice, Lee Murphy & Nicola Wrobel
Torbay Hospital, Torquay, UK
Northern General Hospital, Sheffield, UK
Paul Collini, Cariad Evans & Gary Mills
Liverpool Clinical Trials Centre, University of Liverpool, Liverpool, UK
Marie Connor, Jo Dalton, Chloe Donohue, Carrol Gamble, Michelle Girvan, Sophie Halpin, Janet Harrison, Clare Jackson, Laura Marsh, Stephanie Roberts & Egle Saviciute
Department of Infectious Disease, Imperial College London, London, UK
Graham S. Cooke & Shiranee Sriskandan
St Georges Hospital (Tooting), London, UK
Catherine Cosgrove
Blackpool Victoria Hospital, Blackpool, UK
Jason Cupitt & Joanne Howard
The Royal London Hospital, London, UK
Maria-Teresa Cutino-Moguel
MRC-University of Glasgow Centre for Virus Research, Glasgow, UK
Ana da Silva Filipe, Antonia Y. W. Ho, Sarah E. McDonald, Massimo Palmarini, David L. Robertson, Janet T. Scott & Emma C. Thomson
Salford Royal Hospital, Salford, UK
University Hospital of North Durham, Durham, UK
Chris Dawson
Norfolk and Norwich University Hospital, Norwich, UK
Samir Dervisevic
Intensive Care Unit, Royal Infirmary Edinburgh, Edinburgh, UK
Annemarie B. Docherty & Seán Keating
Institute of Infection, Veterinary and Ecological Sciences, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, UK
Cara Donegan & Rebecca G. Spencer
Salisbury District Hospital, Salisbury, UK
Phil Donnison
National Phenome Centre, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Gonçalo dos Santos Correia, Matthew Lewis, Lynn Maslen, Caroline Sands, Zoltan Takats & Panteleimon Takis
Section of Bioanalytical Chemistry, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Gonçalo dos Santos Correia, Matthew Lewis, Lynn Maslen, Caroline Sands & Panteleimon Takis
Guy’s and St Thomas’, NHS Foundation Trust, London, UK
Sam Douthwaite, Michael MacMahon, Marlies Ostermann & Manu Shankar-Hari
The Royal Oldham Hospital, Oldham, UK
Andrew Drummond
European Genomic Institute for Diabetes, Institut Pasteur de Lille, Lille University Hospital, University of Lille, Lille, France
Marc-Emmanuel Dumas
McGill University and Genome Quebec Innovation Centre, Montreal, Qeubec, Canada
National Infection Service, Public Health England, London, UK
Jake Dunning & Maria Zambon
Hereford Count Hospital, Hereford, UK
Ingrid DuRand
Southampton General Hospital, Southampton, UK
Ahilanadan Dushianthan
Northampton General Hospital, Northampton, UK
Tristan Dyer
University Hospital of Wales, Cardiff, UK
Chrisopher Fegan
University Hospitals Bristol NHS Foundation Trust, Bristol, UK
Liverpool School of Tropical Medicine, Liverpool, UK
Tom Fletcher
Leighton Hospital, Crewe, UK
Duncan Fullerton & Elijah Matovu
Manor Hospital, Walsall, UK
Scunthorpe Hospital, Scunthorpe, UK
Sanjeev Garg
Cambridge University Hospital, Cambridge, UK
Effrossyni Gkrania-Klotsas
West Suffolk NHS Foundation Trust, Bury St Edmunds, UK
Basingstoke and North Hampshire Hospital, Basingstoke, UK
Arthur Goldsmith
North Cumberland Infirmary, Carlisle, UK
Clive Graham
Paediatric Liver, GI and Nutrition Centre and MowatLabs, King’s College Hospital, London, UK
Tassos Grammatikopoulos
Institute of Liver Studies, King’s College London, London, UK
Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK
Christopher A. Green
Department of Molecular and Clinical Cancer Medicine, University of Liverpool, Liverpool, UK
William Greenhalf
Institute for Global Health, University College London, London, UK
Rishi K. Gupta
NIHR Health Protection Research Unit, Institute of Infection, Veterinary and Ecological Sciences, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, UK
Hayley Hardwick, Malcolm G. Semple, Tom Solomon & Lance C. W. Turtle
Warwick Hospital, Warwick, UK
Elaine Hardy
Birmingham Children’s Hospital, Birmingham, UK
Stuart Hartshorn
Nottingham City Hospital, Nottingham, UK
Daniel Harvey
Glangwili Hospital Child Health Section, Carmarthen, UK
Peter Havalda
Alder Hey Children’s Hospital, Liverpool, UK
Daniel B. Hawcutt
Department of Infectious Diseases, Queen Elizabeth University Hospital, Glasgow, UK
Antonia Y. W. Ho
Bronglais General Hospital, Aberystwyth, UK
Maria Hobrok
Worthing Hospital, Worthing, UK
Luke Hodgson
Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Peter W. Horby
Rotheram District General Hospital, Rotheram, UK
Anil Hormis
Virology Reference Department, National Infection Service, Public Health England, Colindale Avenue, London, UK
Samreen Ijaz
Royal Free Hospital, London, UK
Michael Jacobs & Padmasayee Papineni
Homerton Hospital, London, UK
Airedale Hospital, Airedale, UK
Paul Jennings
Basildon Hospital, Basildon, UK
Agilan Kaliappan
The Christie NHS Foundation Trust, Manchester, UK
Vidya Kasipandian
University Hospital Lewisham, London, UK
Stephen Kegg
The Whittington Hospital, London, UK
Michael Kelsey
Southmead Hospital, Bristol, UK
Jason Kendall
Sheffield Childrens Hospital, Sheffield, UK
Caroline Kerrison
Royal United Hospital, Bath, UK
Ian Kerslake
Department of Pharmacology, University of Liverpool, Liverpool, UK
Nuffield Department of Medicine, Peter Medawar Building for Pathogen Research, University of Oxford, Oxford, UK
Paul Klenerman
Translational Gastroenterology Unit, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Public Health Scotland, Edinburgh, UK
Susan Knight, Eva Lahnsteiner & Sarah Tait
Western General Hospital, Edinburgh, UK
Oliver Koch
Southend University Hospital NHS Foundation Trust, Southend-on-Sea, UK
Gouri Koduri
Hinchingbrooke Hospital, Huntingdon, UK
George Koshy & Tamas Leiner
Royal Preston Hospital, Fulwood, UK
Shondipon Laha
University Hospital (Coventry), Coventry, UK
Steven Laird
The Walton Centre, Liverpool, UK
Susan Larkin
ISARIC, Global Support Centre, COVID-19 Clinical Research Resources, Epidemic diseases Research Group, Oxford (ERGO), University of Oxford, Oxford, UK
James Lee & Daniel Plotkin
Centre for Health Informatics, Division of Informatics, Imaging and Data Science, School of Health Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester, UK
Gary Leeming
Hull Royal Infirmary, Hull, UK
Patrick Lillie
Nottingham University Hospitals NHS Trust:, Nottingham, UK
Wei Shen Lim
Darlington Memorial Hospital, Darlington, UK
Queen Elizabeth Hospital (Gateshead), Gateshead, UK
Vanessa Linnett
Warrington Hospital, Warrington, UK
Jeff Little
Bristol Royal Hospital for Children, Bristol, UK
Mark Lyttle
St Mary’s Hospital (Isle of Wight), Isle of Wight, UK
Emily MacNaughton
The Tunbridge Wells Hospital, Royal Tunbridge Wells, UK
Ravish Mankregod
Huddersfield Royal, Huddersfield, UK
Countess of Chester Hospital, Liverpool, UK
Ruth McEwen & Lawrence Wilson
Frimley Park Hospital, Frimley, UK
Manjula Meda
Nuffield Department of Medicine, John Radcliffe Hospital, Oxford, UK
Alexander J. Mentzer
Department of Microbiology/Infectious Diseases, Oxford University Hospitals NHS Foundation Trust, John Radcliffe Hospital, Oxford, UK
MRC Human Genetics Unit, MRC Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK
Alison M. Meynert & Murray Wham
St James University Hospital, Leeds, UK
Jane Minton
Arrowe Park Hospital, Birkenhead, UK
Kavya Mohandas
Great Ormond Street Hospital, London, UK
Royal Shrewsbury Hospital, Shrewsbury, UK
Addenbrookes Hospital, Cambridge, UK
Elinoor Moore
Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK
Shona C. Moore, William A. Paxton & Georgios Pollakis
East Surrey Hospital (Redhill), Redhill, UK
Patrick Morgan
Burton Hospital, Burton, UK
Craig Morris & Tim Reynolds
Peterborough City Hospital, Peterborough, UK
Katherine Mortimore
Kent and Canterbury Hospital, Canterbury, UK
Samuel Moses
Weston Area General Trust, Bristol, UK
Mbiye Mpenge
Bedfordshire Hospital, Bedfordshire, UK
Rohinton Mulla
Glasgow Royal Infirmary, Glasgow, UK
Michael Murphy
Macclesfield General Hospital, Macclesfield, UK
Thapas Nagarajan
Derbyshire Healthcare, Derbyshire, UK
Megan Nagel
Chelsea and Westminster Hospital, London, UK
Mark Nelson & Matthew K. O’Shea
Watford General Hospital, Watford, UK
Lillian Norris & Tom Stambach
EPCC, University of Edinburgh, Edinburgh, UK
Lucy Norris
Section of Biomolecular Medicine, Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, London, UK
Michael Olanipekun
Imperial College Healthcare NHS Trust: London, London, UK
Peter J. M. Openshaw
Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Anthonia Osagie
Prince Philip Hospital, Llanelli, UK
Igor Otahal & Andrew Workman
George Eliot Hospital – Acute Services, Nuneaton, UK
Molecular and Clinical Cancer Medicine, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, UK
Carlo Palmieri
Clatterbridge Cancer Centre NHS Foundation Trust, Liverpool, UK
Kettering General Hospital, Kettering, UK
Selva Panchatsharam
University Hospitals of North Midlands NHS Trust, North Midlands, UK
Danai Papakonstantinou
Russells Hall Hospital, Dudley, UK
Hassan Paraiso
Harefield Hospital, Harefield, UK
Lister Hospital, Lister, UK
Natalie Pattison
Musgrove Park Hospital, Taunton, UK
Justin Pepperell
Kingston Hospital, Kingston, UK
Mark Peters
Queen’s Hospital, Romford, UK
Mandeep Phull
Southport and Formby District General Hospital, Southport, UK
Stefania Pintus
St George’s University of London, London, UK
Tim Planche
King’s College Hospital (Denmark Hill), London, UK
Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, School of Immunology and Microbial Sciences, King’s College London, London, UK
Nicholas Price
Department of Infectious Diseases, Guy’s and St Thomas’ NHS Foundation Trust, London, UK
The Clatterbridge Cancer Centre NHS Foundation, Bebington, UK
David Price
The Great Western Hospital, Swindon, UK
Rachel Prout
Ninewells Hospital, Dundee, UK
Nikolas Rae
Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK
Andrew Rambaut
Poole Hospital NHS Trust, Poole, UK
Henrik Reschreiter
William Harvey Hospital, Ashford, UK
Neil Richardson
King’s Mill Hospital, Sutton-in-Ashfield, UK
Mark Roberts
Liverpool Women’s Hospital, Liverpool, UK
Devender Roberts
Pinderfields Hospital, Wakefield, UK
Alistair Rose
North Devon District Hospital, Barnstaple, UK
Guy Rousseau
Queen Elizabeth Hospital, Birmingham, UK
Tameside General Hospital, Ashton-under-Lyne, UK
Brendan Ryan
City Hospital (Birmingham), Birmingham, UK
Taranprit Saluja
Department of Pediatrics and Virology, St Mary’s Medical School Bldg, Imperial College London, London, UK
Vanessa Sancho-Shimizu
The Newcastle Upon Tyne Hospitals NHS Foundation Trust, Newcastle Upon Tyne, UK
Matthias Schmid
NHS Greater Glasgow and Clyde, Glasgow, UK
Janet T. Scott
Respiratory Medicine, Institute in The Park, University of Liverpool, Alder Hey Children’s Hospital, Liverpool, UK
Malcolm G. Semple
Broomfield Hospital, Broomfield, UK
Stoke Mandeville, UK
Prad Shanmuga
University Hospital of North Tees, Stockton-on-Tees, UK
Anil Sharma
Institute of Translational Medicine, University of, Liverpool, Merseyside, UK
Victoria E. Shaw
Royal Manchester Children’s Hospital, Manchester, UK
Anna Shawcross
New Cross Hospital, Wolverhampton, UK
Jagtur Singh Pooni
Bedford Hospital, Bedford, UK
Jeremy Sizer
Colchester General Hospital, Colchester, UK
Richard Smith
University Hospital Birmingham NHS Foundation Trust, Birmingham, UK
Catherine Snelson & Tony Whitehouse
Walton Centre NHS Foundation Trust, Liverpool, UK
Tom Solomon
Chesterfield Royal Hospital, Calow, UK
Nick Spittle
MRC Centre for Molecular Bacteriology and Infection, Imperial College London, London, UK
Shiranee Sriskandan
Princess Alexandra Hospital, Harlow, UK
Nikki Staines & Shico Visuvanathan
Milton Keynes Hospital, Eaglestone, UK
Richard Stewart
Division of Structural Biology, The Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
David Stuart
Royal Bolton Hopital, Farnworth, UK
Pradeep Subudhi
Department of Medicine, University of Cambridge, Cambridge, UK
Charlotte Summers
Department of Child Life and Health, University of Edinburgh, Edinburgh, UK
Olivia V. Swann
Royal Gwent (Newport), Newport, UK
Tamas Szakmany
The Royal Marsden Hospital (London), London, UK
Kate Tatham
Blood Borne Virus Unit, Virus Reference Department, National Infection Service, Public Health England, London, UK
Richard S. Tedder
Transfusion Microbiology, National Health Service Blood and Transplant, London, UK
Department of Medicine, Imperial College London, London, UK
Queen Victoria Hospital (East Grinstead), East Grinstead, UK
Leeds Teaching Hospitals NHS Trust, Leeds, UK
Robert Thompson
Royal Stoke University Hospital, Stoke-on-Trent, UK
Chris Thompson
Whiston Hospital, Rainhill, UK
Ascanio Tridente
Tropical and Infectious Disease Unit, Royal Liverpool University Hospital, Liverpool, UK
Lance C. W. Turtle
Croydon University Hospital, Thornton Heath, UK
Mary Twagira
Gloucester Royal, Gloucester, UK
Nick Vallotton
West Hertfordshire Teaching Hospitals NHS Trust, Hertfordshire, UK
Rama Vancheeswaran
North Middlesex Hospital, London, UK
Rachel Vincent
Medway Maritime Hospital, Gillingham, UK
Lisa Vincent-Smith
Royal Papworth Hospital Everard, Cambridge, UK
Alan Vuylsteke
Derriford (Plymouth), Plymouth, UK
St Helier Hospital, Sutton, UK
Rachel Wake
Royal Berkshire Hospital, Reading, UK
Andrew Walden
Royal Liverpool Hospital, Liverpool, UK
Ingeborg Welters
Bradford Royal infirmary, Bradford, UK
Paul Whittaker
Central Middlesex, London, UK
Ashley Whittington
Royal Cornwall Hospital (Tresliske), Truro, UK
Meme Wijesinghe
North Bristol NHS Trust, Bristol, UK
Martin Williams
St. Peter’s Hospital, Runnymede, UK
Stephen Winchester
Leicester Royal Infirmary, Leicester, UK
Martin Wiselka
Grantham and District Hospital, Grantham, UK
Adam Wolverson
Aintree University Hospital, Liverpool, UK
Daniel G. Wootton
North Tyneside General Hospital, North Shields, UK
Bryan Yates
Queen Elizabeth Hospital, King’s Lynn, UK
Peter Young
You can also search for this author in PubMed Google Scholar
PHOSP-COVID collaborative group
- Kathryn Abel
- , H. Adamali
- , Davies Adeloye
- , Oluwaseun Adeyemi
- , Rita Adrego
- , Laura Aguilar Jimenez
- , Shanaz Ahmad
- , N. Ahmad Haider
- , Rubina Ahmed
- , Nyarko Ahwireng
- , Mark Ainsworth
- , Asma Alamoudi
- , Mariam Ali
- , M. Aljaroof
- , Louise Allan
- , Richard Allen
- , Lisa Allerton
- , Lynne Allsop
- , Ann Marie Allt
- , Paula Almeida
- , Bashar Al-Sheklly
- , Danny Altmann
- , Maria Alvarez Corral
- , Shannon Amoils
- , David Anderson
- , Charalambos Antoniades
- , Gill Arbane
- , Ava Maria Arias
- , Cherie Armour
- , Lisa Armstrong
- , Natalie Armstrong
- , David Arnold
- , H. Arnold
- , A. Ashish
- , Andrew Ashworth
- , M. Ashworth
- , Shahab Aslani
- , Hosanna Assefa-Kebede
- , Paul Atkin
- , Catherine Atkin
- , Raminder Aul
- , Hnin Aung
- , Liam Austin
- , Cristina Avram
- , Nikos Avramidis
- , Marta Babores
- , Rhiannon Baggott
- , J. Bagshaw
- , David Baguley
- , Elisabeth Bailey
- , J. Kenneth Baillie
- , Steve Bain
- , Majda Bakali
- , E. Baldry
- , Molly Baldwin
- , David Baldwin
- , Clive Ballard
- , Amita Banerjee
- , Dongchun Bang
- , R. E. Barker
- , Laura Barman
- , Perdita Barran
- , Shaney Barratt
- , Fiona Barrett
- , Donna Basire
- , Neil Basu
- , Michelle Bates
- , R. Batterham
- , Helen Baxendale
- , Gabrielle Baxter
- , Hannah Bayes
- , M. Beadsworth
- , Paul Beckett
- , Paul Beirne
- , Murdina Bell
- , Robert Bell
- , Kaytie Bennett
- , Eva Beranova
- , Areti Bermperi
- , Anthony Berridge
- , Colin Berry
- , Sarah Betts
- , Emily Bevan
- , Kamaldeep Bhui
- , Michelle Bingham
- , K. Birchall
- , Lettie Bishop
- , Karen Bisnauthsing
- , John Blaikely
- , Angela Bloss
- , Annette Bolger
- , Charlotte Bolton
- , J. Bonnington
- , A. Botkai
- , Charlotte Bourne
- , Michelle Bourne
- , Kate Bramham
- , Lucy Brear
- , Jonathon Breeze
- , Katie Breeze
- , Andrew Briggs
- , E. Bright
- , Christopher Brightling
- , Simon Brill
- , K. Brindle
- , Lauren Broad
- , Andrew Broadley
- , Claire Brookes
- , Mattew Broome
- , Vanessa Brown
- , Ammani Brown
- , Angela Brown
- , Jeremy Brown
- , Terry Brugha
- , Nigel Brunskill
- , Phil Buckley
- , Anda Bularga
- , Ed Bullmore
- , Jenny Bunker
- , L. Burden
- , Tracy Burdett
- , David Burn
- , John Busby
- , Robyn Butcher
- , Al-Tahoor Butt
- , P. Cairns
- , P. C. Calder
- , Ellen Calvelo
- , H. Carborn
- , Bethany Card
- , Caitlin Carr
- , Liesel Carr
- , G. Carson
- , Penny Carter
- , Anna Casey
- , M. Cassar
- , Jonathon Cavanagh
- , Manish Chablani
- , Trudie Chalder
- , James D. Chalmers
- , Rachel Chambers
- , Flora Chan
- , K. M. Channon
- , Kerry Chapman
- , Amanda Charalambou
- , N. Chaudhuri
- , A. Checkley
- , Yutung Cheng
- , Luke Chetham
- , Caroline Childs
- , Edwin Chilvers
- , H. Chinoy
- , A. Chiribiri
- , K. Chong-James
- , N. Choudhury
- , Gaunab Choudhury
- , Phillip Chowienczyk
- , C. Christie
- , Melanie Chrystal
- , Cameron Clark
- , David Clark
- , Jude Clarke
- , S. Clohisey
- , G. Coakley
- , Zach Coburn
- , S. Coetzee
- , Joby Cole
- , Chris Coleman
- , Florence Conneh
- , David Connell
- , Bronwen Connolly
- , Lynda Connor
- , Amanda Cook
- , Shirley Cooper
- , B. Cooper
- , Josh Cooper
- , Donna Copeland
- , Tracey Cosier
- , Eamon Coughlan
- , Martina Coulding
- , C. Coupland
- , Thelma Craig
- , Daniele Cristiano
- , Michael Crooks
- , Andy Cross
- , Isabel Cruz
- , P. Cullinan
- , D. Cuthbertson
- , Luke Daines
- , Matthhew Dalton
- , Patrick Daly
- , Alison Daniels
- , Joanne Dasgin
- , Anthony David
- , Ffyon Davies
- , Ellie Davies
- , Kim Davies
- , Gareth Davies
- , Gwyneth Davies
- , Melanie Davies
- , Joy Dawson
- , Camilla Dawson
- , Enya Daynes
- , Anthony De Soyza
- , Bill Deakin
- , Andrew Deans
- , Joanne Deery
- , Sylviane Defres
- , Amanda Dell
- , K. Dempsey
- , Emma Denneny
- , J. Dennis
- , Ruvini Dharmagunawardena
- , Nawar Diar-Bakerly
- , Caroline Dickens
- , A. Dipper
- , Sarah Diver
- , Shalin Diwanji
- , Myles Dixon
- , R. Djukanovic
- , Hannah Dobson
- , S. L. Dobson
- , Annemarie B. Docherty
- , A. Donaldson
- , N. Dormand
- , Andrew Dougherty
- , Rachael Dowling
- , Stephen Drain
- , Katharine Draxlbauer
- , Katie Drury
- , Pearl Dulawan
- , A. Dunleavy
- , Sarah Dunn
- , Catherine Dupont
- , Joanne Earley
- , Nicholas Easom
- , Carlos Echevarria
- , Sarah Edwards
- , C. Edwardson
- , Claudia Efstathiou
- , Anne Elliott
- , K. Elliott
- , Yvette Ellis
- , Anne Elmer
- , Omer Elneima
- , Hosni El-Taweel
- , Teriann Evans
- , Ranuromanana Evans
- , Rachael A. Evans
- , Jonathon Evans
- , Cerys Evenden
- , Lynsey Evison
- , Laura Fabbri
- , Sara Fairbairn
- , Alexandra Fairman
- , K. Fallon
- , David Faluyi
- , Clair Favager
- , Tamanah Fayzan
- , James Featherstone
- , T. Felton
- , V. Ferreira
- , Selina Finney
- , J. Finnigan
- , L. Finnigan
- , Helen Fisher
- , S. Fletcher
- , Rachel Flockton
- , Margaret Flynn
- , David Foote
- , Amber Ford
- , D. Forton
- , Eva Fraile
- , C. Francis
- , Richard Francis
- , Susan Francis
- , Anew Frankel
- , Emily Fraser
- , N. French
- , Jonathon Fuld
- , J. Furniss
- , Lucie Garner
- , N. Gautam
- , John Geddes
- , J. George
- , P. George
- , Michael Gibbons
- , Rhyan Gill
- , Mandy Gill
- , L. Gilmour
- , F. Gleeson
- , Jodie Glossop
- , Sarah Glover
- , Nicola Goodman
- , Camelia Goodwin
- , Bibek Gooptu
- , Hussain Gordon
- , T. Gorsuch
- , M. Greatorex
- , Paul Greenhaff
- , William Greenhalf
- , Alan Greenhalgh
- , Neil J. Greening
- , John Greenwood
- , Rebecca Gregory
- , Heidi Gregory
- , D. Grieve
- , Denise Griffin
- , L. Griffiths
- , Anne-Marie Guerdette
- , Beatriz Guillen-Guio
- , Mahitha Gummadi
- , Ayushman Gupta
- , Sambasivarao Gurram
- , Elspeth Guthrie
- , Kate Hadley
- , Ahmed Haggar
- , Kera Hainey
- , Brigid Hairsine
- , Pranab Haldar
- , Lucy Hall
- , Mark Halling-Brown
- , Alyson Hancock
- , Kia Hancock
- , Neil Hanley
- , Sulaimaan Haq
- , Hayley Hardwick
- , Tim Hardy
- , Beverley Hargadon
- , Kate Harrington
- , Edward Harris
- , Victoria C. Harris
- , Ewen Harrison
- , Paul Harrison
- , Nicholas Hart
- , Alice Harvey
- , Matt Harvey
- , M. Harvie
- , L. Haslam
- , Claire Hastie
- , May Havinden-Williams
- , Jenny Hawkes
- , Nancy Hawkings
- , Jill Haworth
- , A. Hayday
- , Matthew Haynes
- , J. Hazeldine
- , Tracy Hazelton
- , Liam Heaney
- , Cheryl Heeley
- , Jonathon Heeney
- , M. Heightman
- , Simon Heller
- , Max Henderson
- , Helen Henson
- , L. Hesselden
- , Melanie Hewitt
- , Victoria Highett
- , T. Hillman
- , Ling-Pei Ho
- , Michaela Hoare
- , Amy Hoare
- , J. Hockridge
- , Philip Hogarth
- , Ailsa Holbourn
- , Sophie Holden
- , L. Holdsworth
- , D. Holgate
- , Maureen Holland
- , Leah Holloway
- , Katie Holmes
- , Megan Holmes
- , B. Holroyd-Hind
- , Anil Hormis
- , Alexander Horsley
- , Akram Hosseini
- , M. Hotopf
- , Linzy Houchen-Wolloff
- , Luke S. Howard
- , Kate Howard
- , Alice Howell
- , E. Hufton
- , Rachel Ann Hughes
- , Joan Hughes
- , Alun Hughes
- , Amy Humphries
- , Nathan Huneke
- , E. Hurditch
- , John Hurst
- , Masud Husain
- , Tracy Hussell
- , John Hutchinson
- , W. Ibrahim
- , Julie Ingham
- , L. Ingram
- , Diana Ionita
- , Karen Isaacs
- , Khalida Ismail
- , T. Jackson
- , Joseph Jacob
- , W. Y. James
- , Claire Jarman
- , Ian Jarrold
- , Hannah Jarvis
- , Roman Jastrub
- , Bhagy Jayaraman
- , Gisli Jenkins
- , P. Jezzard
- , Kasim Jiwa
- , C. Johnson
- , Simon Johnson
- , Desmond Johnston
- , Caroline Jolley
- , Ian Jones
- , Heather Jones
- , Mark Jones
- , Don Jones
- , Sherly Jose
- , Thomas Kabir
- , G. Kaltsakas
- , Vicky Kamwa
- , N. Kanellakis
- , Sabina Kaprowska
- , Zunaira Kausar
- , Natalie Keenan
- , Steven Kerr
- , Helen Kerslake
- , Angela Key
- , Fasih Khan
- , Kamlesh Khunti
- , Susan Kilroy
- , Bernie King
- , Clara King
- , Lucy Kingham
- , Jill Kirk
- , Paaig Kitterick
- , Paul Klenerman
- , Lucy Knibbs
- , Sean Knight
- , Abigail Knighton
- , Onn Min Kon
- , Samantha Kon
- , Ania Korszun
- , Ivan Koychev
- , Claire Kurasz
- , Prathiba Kurupati
- , Hanan Lamlum
- , G. Landers
- , Claudia Langenberg
- , Lara Lavelle-Langham
- , Allan Lawrie
- , Cathy Lawson
- , Claire Lawson
- , Alison Layton
- , Olivia C. Leavy
- , Ju Hee Lee
- , Elvina Lee
- , Karen Leitch
- , Rebecca Lenagh
- , Victoria Lewis
- , Joanne Lewis
- , Keir Lewis
- , N. Lewis-Burke
- , Felicity Liew
- , Tessa Light
- , Liz Lightstone
- , W. Lilaonitkul
- , S. Linford
- , Anne Lingford-Hughes
- , M. Lipman
- , Kamal Liyanage
- , Arwel Lloyd
- , Nazir I. Lone
- , Ronda Loosley
- , Janet Lord
- , Harpreet Lota
- , Wayne Lovegrove
- , Daniel Lozano-Rojas
- , Alice Lucey
- , Gardiner Lucy
- , E. Lukaschuk
- , Alison Lye
- , Ceri Lynch
- , S. MacDonald
- , G. MacGowan
- , Irene Macharia
- , J. Mackie
- , L. Macliver
- , S. Madathil
- , Gladys Madzamba
- , Nick Magee
- , Murphy Magtoto
- , N. Majeed
- , Flora Malein
- , Georgia Mallison
- , William Man
- , S. Mandal
- , K. Mangion
- , C. Manisty
- , R. Manley
- , Katherine March
- , Stefan Marciniak
- , Philip Marino
- , Myril Mariveles
- , Michael Marks
- , Elizabeth Marouzet
- , Sophie Marsh
- , M. Marshall
- , B. Marshall
- , Jane Martin
- , Adrian Martineau
- , L. M. Martinez
- , Nick Maskell
- , Darwin Matila
- , Wadzanai Matimba-Mupaya
- , Laura Matthews
- , Angeline Mbuyisa
- , Steve McAdoo
- , Hamish McAllister-Williams
- , Paul McArdle
- , Anne McArdle
- , Danny McAulay
- , Hamish J. C. McAuley
- , Gerry McCann
- , W. McCormick
- , Jacqueline McCormick
- , P. McCourt
- , Celeste McCracken
- , Lorcan McGarvey
- , Jade McGinness
- , K. McGlynn
- , Andrew McGovern
- , Heather McGuinness
- , I. B. McInnes
- , Jerome McIntosh
- , Emma McIvor
- , Katherine McIvor
- , Laura McLeavey
- , Aisling McMahon
- , Michael McMahon
- , L. McMorrow
- , Teresa Mcnally
- , M. McNarry
- , J. McNeill
- , Alison McQueen
- , H. McShane
- , Chloe Mears
- , Clare Megson
- , Sharon Megson
- , J. Meiring
- , Lucy Melling
- , Mark Mencias
- , Daniel Menzies
- , Marta Merida Morillas
- , Alice Michael
- , Benedict Michael
- , C. A. Miller
- , Lea Milligan
- , Nicholas Mills
- , Clare Mills
- , George Mills
- , L. Milner
- , Jane Mitchell
- , Abdelrahman Mohamed
- , Noura Mohamed
- , S. Mohammed
- , Philip Molyneaux
- , Will Monteiro
- , Silvia Moriera
- , Anna Morley
- , Leigh Morrison
- , Richard Morriss
- , A. Morrow
- , Paul Moss
- , Alistair Moss
- , K. Motohashi
- , N. Msimanga
- , Elizabeta Mukaetova-Ladinska
- , Unber Munawar
- , Jennifer Murira
- , Uttam Nanda
- , Heeah Nassa
- , Mariam Nasseri
- , Rashmita Nathu
- , Aoife Neal
- , Robert Needham
- , Paula Neill
- , Stefan Neubauer
- , D. E. Newby
- , Helen Newell
- , J. Newman
- , Tom Newman
- , Alex Newton-Cox
- , T. E. Nichols
- , Tim Nicholson
- , Christos Nicolaou
- , Debby Nicoll
- , Athanasios Nikolaidis
- , C. Nikolaidou
- , C. M. Nolan
- , Matthew Noonan
- , C. Norman
- , Petr Novotny
- , Kimon Ntotsis
- , Jose Nunag
- , Lorenza Nwafor
- , Uchechi Nwanguma
- , Joseph Nyaboko
- , Linda O’Brien
- , C. O’Brien
- , Natasha Odell
- , Kate O’Donnell
- , Godwin Ogbole
- , Olaoluwa Olaosebikan
- , Catherine Oliver
- , Zohra Omar
- , Peter J. M. Openshaw
- , D. P. O’Regan
- , Lorna Orriss-Dib
- , Lynn Osborne
- , Rebecca Osbourne
- , Marlies Ostermann
- , Charlotte Overton
- , Jamie Pack
- , Edmund Pacpaco
- , Stella-Maria Paddick
- , Sharon Painter
- , Erola Pairo-Castineira
- , Ashkan Pakzad
- , Sue Palmer
- , Padmasayee Papineni
- , K. Paques
- , Kerry Paradowski
- , Manish Pareek
- , Dhruv Parekh
- , H. Parfrey
- , Carmen Pariante
- , S. Parker
- , M. Parkes
- , J. Parmar
- , Sheetal Patale
- , Manish Patel
- , Suhani Patel
- , Dibya Pattenadk
- , M. Pavlides
- , Sheila Payne
- , Lorraine Pearce
- , John Pearl
- , Dan Peckham
- , Jessica Pendlebury
- , Yanchun Peng
- , Chris Pennington
- , Ida Peralta
- , Emma Perkins
- , Z. Peterkin
- , Tunde Peto
- , Nayia Petousi
- , John Petrie
- , Paul Pfeffer
- , Janet Phipps
- , S. Piechnik
- , John Pimm
- , Karen Piper Hanley
- , Riinu Pius
- , Hannah Plant
- , Tatiana Plekhanova
- , Megan Plowright
- , Krisnah Poinasamy
- , Oliver Polgar
- , Julie Porter
- , Joanna Porter
- , Sofiya Portukhay
- , Natassia Powell
- , A. Prabhu
- , James Pratt
- , Andrea Price
- , Claire Price
- , Carly Price
- , Anne Prickett
- , I. Propescu
- , J. Propescu
- , Sabrina Prosper
- , S. Pugmire
- , Sheena Quaid
- , Jackie Quigley
- , Jennifer K. Quint
- , H. Qureshi
- , I. N. Qureshi
- , K. Radhakrishnan
- , Najib Rahman
- , Markus Ralser
- , Betty Raman
- , Hazel Ramos
- , Albert Ramos
- , Jade Rangeley
- , Bojidar Rangelov
- , Liz Ratcliffe
- , Phillip Ravencroft
- , Konrad Rawlik
- , Anne Reddington
- , Heidi Redfearn
- , Dawn Redwood
- , Annabel Reed
- , Meryl Rees
- , Tabitha Rees
- , Karen Regan
- , Will Reynolds
- , Carla Ribeiro
- , A. Richards
- , Emma Richardson
- , M. Richardson
- , Pilar Rivera-Ortega
- , K. Roberts
- , Elizabeth Robertson
- , Leanne Robinson
- , Emma Robinson
- , Lisa Roche
- , C. Roddis
- , J. Rodger
- , Natalie Rogers
- , Gavin Ross
- , Alexandra Ross
- , Jennifer Rossdale
- , Anthony Rostron
- , Anna Rowe
- , J. Rowland
- , M. J. Rowland
- , A. Rowland
- , Sarah L. Rowland-Jones
- , Maura Roy
- , Igor Rudan
- , Richard Russell
- , Emily Russell
- , Gwen Saalmink
- , Ramsey Sabit
- , Beth Sage
- , T. Samakomva
- , Nilesh Samani
- , A. A. Samat
- , Claire Sampson
- , Katherine Samuel
- , Reena Samuel
- , Z. B. Sanders
- , Amy Sanderson
- , Elizabeth Sapey
- , Dinesh Saralaya
- , Jack Sargant
- , Carol Sarginson
- , Naveed Sattar
- , Kathryn Saunders
- , Peter Saunders
- , Ruth Saunders
- , Laura Saunders
- , Heather Savill
- , Avan Sayer
- , J. Schronce
- , William Schwaeble
- , Janet Scott
- , Kathryn Scott
- , Nick Selby
- , Malcolm G. Semple
- , Marco Sereno
- , Terri Ann Sewell
- , Kamini Shah
- , Ajay Shah
- , Manu Shankar-Hari
- , M. Sharma
- , Claire Sharpe
- , Michael Sharpe
- , Sharlene Shashaa
- , Alison Shaw
- , Victoria Shaw
- , Karen Shaw
- , Aziz Sheikh
- , Sarah Shelton
- , Liz Shenton
- , K. Shevket
- , Aarti Shikotra
- , Sulman Siddique
- , Salman Siddiqui
- , J. Sidebottom
- , Louise Sigfrid
- , Gemma Simons
- , Neil Simpson
- , John Simpson
- , Ananga Singapuri
- , Suver Singh
- , Claire Singh
- , Sally Singh
- , D. Sissons
- , J. Skeemer
- , Katie Slack
- , David Smith
- , Nikki Smith
- , Andrew Smith
- , Jacqui Smith
- , Laurie Smith
- , Susan Smith
- , M. Soares
- , Teresa Solano
- , Reanne Solly
- , A. R. Solstice
- , Tracy Soulsby
- , David Southern
- , D. Sowter
- , Mark Spears
- , Lisa Spencer
- , Fabio Speranza
- , Louise Stadon
- , Stefan Stanel
- , R. Steeds
- , N. Steele
- , Mike Steiner
- , David Stensel
- , G. Stephens
- , Lorraine Stephenson
- , Iain Stewart
- , R. Stimpson
- , Sue Stockdale
- , J. Stockley
- , Wendy Stoker
- , Roisin Stone
- , Will Storrar
- , Andrew Storrie
- , Kim Storton
- , E. Stringer
- , Sophia Strong-Sheldrake
- , Natalie Stroud
- , Christian Subbe
- , Catherine Sudlow
- , Zehra Suleiman
- , Charlotte Summers
- , C. Summersgill
- , Debbie Sutherland
- , D. L. Sykes
- , Nick Talbot
- , Ai Lyn Tan
- , Lawrence Tarusan
- , Vera Tavoukjian
- , Jessica Taylor
- , Abigail Taylor
- , Chris Taylor
- , John Paul Taylor
- , Amelie Te
- , Caroline Tee
- , J. Teixeira
- , Helen Tench
- , Sarah Terry
- , Susannah Thackray-Nocera
- , Favas Thaivalappil
- , David Thickett
- , David Thomas
- , S. Thomas
- , Caradog Thomas
- , Andrew Thomas
- , T. Thomas-Woods
- , A. A. Roger Thompson
- , Tamika Thompson
- , T. Thornton
- , Matthew Thorpe
- , Ryan S. Thwaites
- , Jo Tilley
- , N. Tinker
- , Gerlynn Tiongson
- , Martin Tobin
- , Johanne Tomlinson
- , Mark Toshner
- , T. Treibel
- , K. A. Tripp
- , Drupad Trivedi
- , E. M. Tunnicliffe
- , Alison Turnbull
- , Kim Turner
- , Sarah Turner
- , Victoria Turner
- , E. Turner
- , Sharon Turney
- , Lance Turtle
- , Helena Turton
- , Jacinta Ugoji
- , R. Ugwuoke
- , Rachel Upthegrove
- , Jonathon Valabhji
- , Maximina Ventura
- , Joanne Vere
- , Carinna Vickers
- , Ben Vinson
- , Ioannis Vogiatzis
- , Elaine Wade
- , Phillip Wade
- , Louise V. Wain
- , Tania Wainwright
- , Lilian Wajero
- , Sinead Walder
- , Samantha Walker
- , S. Walker
- , Tim Wallis
- , Sarah Walmsley
- , Simon Walsh
- , J. A. Walsh
- , Louise Warburton
- , T. J. C. Ward
- , Katie Warwick
- , Helen Wassall
- , Samuel Waterson
- , L. Watson
- , Ekaterina Watson
- , James Watson
- , M. Webster
- , J. Weir McCall
- , Carly Welch
- , Simon Wessely
- , Sophie West
- , Heather Weston
- , Helen Wheeler
- , Sonia White
- , Victoria Whitehead
- , J. Whitney
- , S. Whittaker
- , Beverley Whittam
- , V. Whitworth
- , Andrew Wight
- , James Wild
- , Martin Wilkins
- , Dan Wilkinson
- , Nick Williams
- , N. Williams
- , B. Williams
- , Jenny Williams
- , S. A. Williams-Howard
- , Michelle Willicombe
- , Gemma Willis
- , James Willoughby
- , Ann Wilson
- , Imogen Wilson
- , Daisy Wilson
- , Nicola Window
- , M. Witham
- , Rebecca Wolf-Roberts
- , Chloe Wood
- , F. Woodhead
- , Janet Woods
- , Dan Wootton
- , J. Wormleighton
- , J. Worsley
- , David Wraith
- , Caroline Wrey Brown
- , C. Wright
- , S. Wright
- , Louise Wright
- , Inez Wynter
- , Moucheng Xu
- , Najira Yasmin
- , S. Yasmin
- , Tom Yates
- , Kay Por Yip
- , Susan Young
- , Bob Young
- , A. J. Yousuf
- , Amira Zawia
- , Lisa Zeidan
- , Bang Zhao
- , Bang Zheng
- & O. Zongo
- , Daniel Agranoff
- , Ken Agwuh
- , Katie A. Ahmed
- , Dhiraj Ail
- , Erin L. Aldera
- , Ana Alegria
- , Beatrice Alex
- , Sam Allen
- , Petros Andrikopoulos
- , Brian Angus
- , Jane A. Armstrong
- , Abdul Ashish
- , Milton Ashworth
- , Innocent G. Asiimwe
- , Dougal Atkinson
- , Benjamin Bach
- , Siddharth Bakshi
- , Wendy S. Barclay
- , Shahedal Bari
- , Gavin Barlow
- , Samantha L. Barlow
- , Stella Barnass
- , Nicholas Barrett
- , Christopher Bassford
- , Sneha Basude
- , David Baxter
- , Michael Beadsworth
- , Jolanta Bernatoniene
- , John Berridge
- , Nicola Best
- , Debby Bogaert
- , Laura Booth
- , Pieter Bothma
- , Benjamin Brennan
- , Robin Brittain-Long
- , Katie Bullock
- , Naomi Bulteel
- , Tom Burden
- , Andrew Burtenshaw
- , Nicola Carlucci
- , Gail Carson
- , Vikki Caruth
- , Emily Cass
- , Benjamin W. A. Catterall
- , David Chadwick
- , Duncan Chambler
- , Meera Chand
- , Kanta Chechi
- , Nigel Chee
- , Jenny Child
- , Srikanth Chukkambotla
- , Richard Clark
- , Tom Clark
- , Jordan J. Clark
- , Emily A. Clarke
- , Sara Clohisey
- , Sarah Cole
- , Paul Collini
- , Marie Connor
- , Graham S. Cooke
- , Louise Cooper
- , Catherine Cosgrove
- , Audrey Coutts
- , Helen Cox
- , Jason Cupitt
- , Maria-Teresa Cutino-Moguel
- , Ana da Silva Filipe
- , Jo Dalton
- , Paul Dark
- , Christopher Davis
- , Chris Dawson
- , Thushan de Silva
- , Samir Dervisevic
- , Oslem Dincarslan
- , Alejandra Doce Carracedo
- , Cara Donegan
- , Lorna Donelly
- , Phil Donnison
- , Chloe Donohue
- , Gonçalo dos Santos Correia
- , Sam Douthwaite
- , Thomas M. Drake
- , Andrew Drummond
- , Marc-Emmanuel Dumas
- , Chris Dunn
- , Jake Dunning
- , Ingrid DuRand
- , Ahilanadan Dushianthan
- , Tristan Dyer
- , Philip Dyer
- , Angela Elliott
- , Cariad Evans
- , Anthony Evans
- , Chi Eziefula
- , Cameron J. Fairfield
- , Angie Fawkes
- , Chrisopher Fegan
- , Lorna Finch
- , Adam Finn
- , Lewis W. S. Fisher
- , Lisa Flaherty
- , Tom Fletcher
- , Terry Foster
- , Duncan Fullerton
- , Carrol Gamble
- , Isabel Garcia-Dorival
- , Atul Garg
- , Sanjeev Garg
- , Tammy Gilchrist
- , Michelle Girvan
- , Effrossyni Gkrania-Klotsas
- , Jo Godden
- , Arthur Goldsmith
- , Clive Graham
- , Tassos Grammatikopoulos
- , Christopher A. Green
- , Julian Griffin
- , Fiona Griffiths
- , Philip Gunning
- , Rishi K. Gupta
- , Katarzyna Hafezi
- , Sophie Halpin
- , Elaine Hardy
- , Ewen M. Harrison
- , Janet Harrison
- , Catherine Hartley
- , Stuart Hartshorn
- , Daniel Harvey
- , Peter Havalda
- , Daniel B. Hawcutt
- , Ross Hendry
- , Antonia Y. W. Ho
- , Maria Hobrok
- , Luke Hodgson
- , Karl Holden
- , Anthony Holmes
- , Peter W. Horby
- , Joanne Howard
- , Samreen Ijaz
- , Clare Jackson
- , Michael Jacobs
- , Susan Jain
- , Paul Jennings
- , Rebecca L. Jensen
- , Christopher B. Jones
- , Trevor R. Jones
- , Agilan Kaliappan
- , Vidya Kasipandian
- , Seán Keating
- , Stephen Kegg
- , Michael Kelsey
- , Jason Kendall
- , Caroline Kerrison
- , Ian Kerslake
- , Shadia Khandaker
- , Katharine King
- , Robyn T. Kiy
- , Stephen R. Knight
- , Susan Knight
- , Oliver Koch
- , Gouri Koduri
- , George Koshy
- , Chrysa Koukorava
- , Shondipon Laha
- , Eva Lahnsteiner
- , Steven Laird
- , Annette Lake
- , Suzannah Lant
- , Susan Larkin
- , Diane Latawiec
- , Andrew Law
- , James Lee
- , Gary Leeming
- , Daniella Lefteri
- , Tamas Leiner
- , Lauren Lett
- , Matthew Lewis
- , Sonia Liggi
- , Patrick Lillie
- , Wei Shen Lim
- , James Limb
- , Vanessa Linnett
- , Jeff Little
- , Lucia A. Livoti
- , Mark Lyttle
- , Louise MacGillivray
- , Alan Maclean
- , Michael MacMahon
- , Emily MacNaughton
- , Maria Mancini
- , Ravish Mankregod
- , Laura Marsh
- , Lynn Maslen
- , Hannah Massey
- , Huw Masson
- , Elijah Matovu
- , Nicole Maziere
- , Sarah McCafferty
- , Katherine McCullough
- , Sarah E. McDonald
- , Sarah McDonald
- , Laurence McEvoy
- , Ruth McEwen
- , John McLauchlan
- , Kenneth A. Mclean
- , Manjula Meda
- , Alexander J. Mentzer
- , Laura Merson
- , Soeren Metelmann
- , Alison M. Meynert
- , Nahida S. Miah
- , Joanna Middleton
- , Gary Mills
- , Jane Minton
- , Joyce Mitchell
- , Kavya Mohandas
- , James Moon
- , Elinoor Moore
- , Shona C. Moore
- , Patrick Morgan
- , Kirstie Morrice
- , Craig Morris
- , Katherine Mortimore
- , Samuel Moses
- , Mbiye Mpenge
- , Rohinton Mulla
- , Derek Murphy
- , Lee Murphy
- , Michael Murphy
- , Ellen G. Murphy
- , Thapas Nagarajan
- , Megan Nagel
- , Mark Nelson
- , Lisa Norman
- , Lillian Norris
- , Lucy Norris
- , Mahdad Noursadeghi
- , Michael Olanipekun
- , Wilna Oosthuyzen
- , Anthonia Osagie
- , Matthew K. O’Shea
- , Igor Otahal
- , Mark Pais
- , Massimo Palmarini
- , Carlo Palmieri
- , Selva Panchatsharam
- , Danai Papakonstantinou
- , Hassan Paraiso
- , Brij Patel
- , Natalie Pattison
- , William A. Paxton
- , Rebekah Penrice-Randal
- , Justin Pepperell
- , Mark Peters
- , Mandeep Phull
- , Jack Pilgrim
- , Stefania Pintus
- , Tim Planche
- , Daniel Plotkin
- , Georgios Pollakis
- , Frank Post
- , Nicholas Price
- , David Price
- , Tessa Prince
- , Rachel Prout
- , Nikolas Rae
- , Andrew Rambaut
- , Henrik Reschreiter
- , Tim Reynolds
- , Neil Richardson
- , P. Matthew Ridley
- , Mark Roberts
- , Stephanie Roberts
- , Devender Roberts
- , David L. Robertson
- , Alistair Rose
- , Guy Rousseau
- , Bobby Ruge
- , Clark D. Russell
- , Brendan Ryan
- , Debby Sales
- , Taranprit Saluja
- , Vanessa Sancho-Shimizu
- , Caroline Sands
- , Egle Saviciute
- , Matthias Schmid
- , Janet T. Scott
- , James Scott-Brown
- , Aarti Shah
- , Prad Shanmuga
- , Anil Sharma
- , Catherine A. Shaw
- , Victoria E. Shaw
- , Anna Shawcross
- , Rebecca K. Shears
- , Jagtur Singh Pooni
- , Jeremy Sizer
- , Benjamin Small
- , Richard Smith
- , Catherine Snelson
- , Tom Solomon
- , Rebecca G. Spencer
- , Nick Spittle
- , Shiranee Sriskandan
- , Nikki Staines
- , Tom Stambach
- , Richard Stewart
- , David Stuart
- , Krishanthi S. Subramaniam
- , Pradeep Subudhi
- , Olivia V. Swann
- , Tamas Szakmany
- , Agnieska Szemiel
- , Aislynn Taggart
- , Sarah Tait
- , Zoltan Takats
- , Panteleimon Takis
- , Jolanta Tanianis-Hughes
- , Kate Tatham
- , Richard S. Tedder
- , Jo Thomas
- , Jordan Thomas
- , Robert Thompson
- , Chris Thompson
- , Emma C. Thomson
- , Ascanio Tridente
- , Erwan Trochu
- , Darell Tupper-Carey
- , Lance C. W. Turtle
- , Mary Twagira
- , Nick Vallotton
- , Libby van Tonder
- , Rama Vancheeswaran
- , Rachel Vincent
- , Lisa Vincent-Smith
- , Shico Visuvanathan
- , Alan Vuylsteke
- , Sam Waddy
- , Rachel Wake
- , Andrew Walden
- , Ingeborg Welters
- , Murray Wham
- , Tony Whitehouse
- , Paul Whittaker
- , Ashley Whittington
- , Meme Wijesinghe
- , Eve Wilcock
- , Martin Williams
- , Lawrence Wilson
- , Stephen Winchester
- , Martin Wiselka
- , Adam Wolverson
- , Daniel G. Wootton
- , Andrew Workman
- , Nicola Wrobel
- , Bryan Yates
- , Peter Young
- , Maria Zambon
- & J. Eunice Zhang
Contributions
F.L. recruited participants, acquired clinical samples, analyzed and interpreted data and cowrote the manuscript, including all drafting and revisions. C.E. analyzed and interpreted data and cowrote this manuscript, including all drafting and revisions. S.F. and M.R. supported the analysis and interpretation of data as well as drafting and revisions. D.S., J.K.S., S.C.M., S.A., N.M., J.N., C.K., O.C.L., O.E., H.J.C.M., A. Shikotra, A. Singapuri, M.S., V.C.H., M.T., N.J.G., N.I.L. and C.C. contributed to acquisition of data underlying this study. L.H.-W., A.A.R.T., S.L.R.-J., L.S.H., O.M.K., D.G.W., T.I.d.S. and A. Ho made substantial contributions to conception/design and implementation of this work and/or acquisition of clinical samples for this work. They have supported drafting and revisions of the manuscript. E.M.H., J.K.Q. and A.B.D. made substantial contributions to the study design as well as data access, linkage and analysis. They have supported drafting and revisions of this work. J.D.C., L.-P.H., A. Horsley, B.R., K.P., M.M. and W.G. made substantial contributions to the conception and design of this work and have supported drafting and revisions of this work. J.K.B. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, has made substantial contributions to conception and design of this work and has supported drafting and revisions of this work. M.G.S. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, sponsor/protocol chief investigator, has made substantial contributions to conception and design of this work and has supported drafting and revisions of this work. R.A.E. and L.V.W. are co-leads of PHOSP-COVID, made substantial contributions to conception and design of this work, the acquisition and analysis of data, and have supported drafting and revisions of this work. C.B. is the chief investigator of PHOSP-COVID and has made substantial contributions to conception and design of this work. R.S.T. and L.T. made substantial contributions to the acquisition, analysis and interpretation of the data underlying this study and have contributed to drafting and revisions of this work. P.J.M.O. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, sponsor/protocol chief investigator and has made substantial contributions to conception and design of this work. R.S.T. and P.J.M.O. have also made key contributions to interpretation of data and have co-written this manuscript. All authors have read and approve the final version to be published. All authors agree to accountability for all aspects of this work. All investigators within ISARIC4C and the PHOSP-COVID consortia have made substantial contributions to the conception or design of this study and/or acquisition of data for this study. The full list of authors within these groups is available in Supplementary Information .
Corresponding authors
Correspondence to Ryan S. Thwaites or Peter J. M. Openshaw .
Ethics declarations
Competing interests.
F.L., C.E., D.S., J.K.S., S.C.M., C.D., C.K., N.M., L.N., E.M.H., A.B.D., J.K.Q., L.-P.H., K.P., L.S.H., O.M.K., S.F., T.I.d.S., D.G.W., R.S.T. and J.K.B. have no conflicts of interest. A.A.R.T. receives speaker fees and support to attend meetings from Janssen Pharmaceuticals. S.L.R.-J. is on the data safety monitoring board for Bexero trial in HIV+ adults in Kenya. J.D.C. is the deputy chief editor of the European Respiratory Journal and receives consulting fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GSK, Insmed, Janssen, Novartis, Pfizer and Zambon. A. Horsley is deputy chair of NIHR Translational Research Collaboration (unpaid role). B.R. receives honoraria from Axcella therapeutics. R.A.E. is co-lead of PHOSP-COVID and receives fees from AstraZenaca/Evidera for consultancy on LC and from AstraZenaca for consultancy on digital health. R.A.E. has received speaker fees from Boehringer in June 2021 and has held a role as European Respiratory Society Assembly 01.02 Pulmonary Rehabilitation secretary. R.A.E. is on the American Thoracic Society Pulmonary Rehabilitation Assembly program committee. L.V.W. also receives funding from Orion pharma and GSK and holds contracts with Genentech and AstraZenaca. L.V.W. has received consulting fees from Galapagos and Boehringer, is on the data advisory board for Galapagos and is Associate Editor for the European Respiratory Journal . A. Ho is a member of NIHR Urgent Public Health Group (June 2020–March 2021). M.M. is an applicant on the PHOSP study funded by NIHR/DHSC. M.G.S. acts as an independent external and nonremunerated member of Pfizer’s External Data Monitoring Committee for their mRNA vaccine program(s), is Chair of Infectious Disease Scientific Advisory Board of Integrum Scientific LLC, and is director of MedEx Solutions Ltd. and majority owner of MedEx Solutions Ltd. and minority owner of Integrum Scientific LLC. M.G.S.’s institution has been in receipt of gifts from Chiesi Farmaceutici S.p.A. of Clinical Trial Investigational Medicinal Product without encumbrance and distribution of same to trial sites. M.G.S. is a nonrenumerated member of HMG UK New Emerging Respiratory Virus Threats Advisory Group and has previously been a nonrenumerated member of the Scientific Advisory Group for Emergencies (SAGE). C.B. has received consulting fees and/or grants from GSK, AstraZeneca, Genentech, Roche, Novartis, Sanofi, Regeneron, Chiesi, Mologic and 4DPharma. L.T. has received consulting fees from MHRA, AstraZeneca and Synairgen and speakers’ fees from Eisai Ltd., and support for conference attendance from AstraZeneca. L.T. has a patent pending with ZikaVac. P.J.M.O. reports grants from the EU Innovative Medicines Initiative 2 Joint Undertaking during the submitted work; grants from UK Medical Research Council, GSK, Wellcome Trust, EU Innovative Medicines Initiative, UK National Institute for Health Research and UK Research and Innovation–Department for Business, Energy and Industrial Strategy; and personal fees from Pfizer, Janssen and Seqirus, outside the submitted work.
Peer review
Peer review information.
Nature Immunology thanks Ziyad Al-Aly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ioana Staicu was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 penalized logistic regression performance..
Graphs show classification error and Area under curve (AUC) from the 50 repeats tenfold nested cross-validation used to optimise and assess the performance of PLR testing associations with each LC outcome relative to Recovered (n = 233): Cardio_Resp (n = 398), Fatigue (n = 384), Anxiety/Depression (n = 202), GI (n = 132), ( e ) Cognitive (n = 6). The distributions of classification error and area under curve (AUC) from the nested cross-validation are shown. Box plot centre line represents the Median and boundaries of the box represent interquartile range (IQR), the whisker length represent 1.5xIQR.
Extended Data Fig. 2 Associations with long COVID symptoms in full study cohort.
( a ) Fibrinogen levels at 6 months were compared between pooled LC cases (n = 295) and Recovered (n = 233) and between the Cognitive group (n = 41) and Recovered (n = 233). Box plot centre line represent the Median and boundaries of the box represent interquartile range (IQR), the whisker length represents 1.5xIQR, any outliers beyond the whisker range are shown as individual dots. Median differences were compared using two-sided Wilcoxon signed-rank test *= p < 0·05, **= p < 0·01, ***= p < 0·001, ****= p < 0·0001. Unadjusted p-values are reported. b ) Distribution of time from COVID-19 hospitalisation at sample collection applying CDC and NICE definitions of LC (n = 719) ( c ) Upset plot of symptom groups. Horizontal coloured bars represent the number of patients in each symptom group: Cardiorespiratory (Cardio_Resp), Fatigue, Cognitive, Gastrointestinal (GI) and Anxiety/Depression (Anx_Dep). Vertical black bars represent the number of patients in each symptom combination group. To prevent patient identification, where less than 5 patients belong to a combination group, this has been represented as ‘<5’. The Recovered group (n = 250) were used as controls. Forest plots show Olink protein concentrations (NPX) associated with ( d ) Cardio_Resp (n = 398), ( e ) Fatigue (n = 342), ( f ) Anx_Dep (n = 219), ( g ) GI (n = 134), and ( h ) Cognitive (n = 65). Error bars represent the median accuracy of the model.
Extended Data Fig. 3 Validation of olink measurements using conventional assays in plasma.
Olink measured protein (NPX) were compared to chemiluminescence assays (ECL or ELISA, log2[pg/mL]) to validate our findings, where contemporaneously collected plasma samples were available (n = 58). Results from key mediators associated with LC groups were validated: CSF3, IL1R2, IL2, IL3RA, TNFa, TFF2. R = spearman rank correlation coefficient and shaded areas indicated the 95% confidence interval. Samples that fell below the lower limit of detection for a given assay were excluded and the ‘n’ value on each panel indicates the number of samples above this limit.
Extended Data Fig. 4 Univariate analysis of proteins associated with each symptom.
Olink measured plasma protein levels (NPX) compared between LC groups (Cardio_Resp, n = 398, Fatigue n = 384, Anxiety/Depression, n = 202, GI, n = 132 and Cognitive, n = 60) and Recovered (n = 233). Proteins identified by PLR were compared between groups. Median differences were compared using two-sided Wilcoxon signed-rank test. * = p < 0·05, ** = p < 0·01, *** = p < 0·001, ****= p < 0·0001 after FDR adjustment. Box plot centre line represent the Median and boundaries of the box represent interquartile range (IQR), the whisker length represents 1.5xIQR, any outliers beyond the whisker range are shown as individual dots.
Extended Data Fig. 5 Unadjusted Penalised Logistic Regression.
Olink measured proteins (NPX) and their association with Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65). Forest plots show odds of each LC outcome vs Recovered (n = 233), using PLR without adjusting for clinical co-variates. Error bars represent the median accuracy of the model.
Extended Data Fig. 6 Partial Least Squares analysis.
Olink measured proteins (NPX) and their association with Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups. Forest plots show odds of LC outcome vs Recovered (n = 233), using PLS analysis. Error bars represent the standard error of the coefficient estimate.
Extended Data Fig. 7 Network analysis centrality.
Each graph shows the centrality score for each Olink measured protein (NPX) found to have significant associations with other proteins that were elevated in the Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups relative to Recovered (n = 233).
Extended Data Fig. 8 Inflammation in men and women with long COVID.
Olink measured plasma protein levels (NPX) between men and women with symptoms, divided by age (<50 or >=50years): (a) shows IL1R2 and MATN2 in the Anxiety/Depression group (<50 n = 55, >=50 n = 133), (b) shows CTSO and NFASC in the Cognitive group (<50 n = 11, >=50 n = 50). Median values were compared between men and women using two-sided Wilcoxon signed-rank test. Box plot centre line represent the Median and boundaries represent interquartile range (IQR), the whisker length represents 1.5xIQR.
Extended Data Fig. 9 Inflammation in the upper respiratory tract.
Nasal cytokines measured by immunoassay in the CardioResp Group (n = 29) and Recovered (n = 31): ( a ) shows IL1a, IL1b, IL-6, APO-2, TGFa, TFF2. Median differences were compared using two-sided Wilcoxon signed-rank test. Box plot centre line represents the Median and boundaries of the box represent interquartile range (IQR), the whisker length represent 1.5xIQR. ( b ) Shows cytokines measured by immunoassay in paired plasma and nasal (n = 70). Correlations between IL1a, IL1b, IL-6, APO-2, TGFa and TFF2 in nasal and plasma samples were compared using Spearman’s rank correlation coefficient ( R ). Shaded areas indicated the 95% confidence interval of R.
Extended Data Fig. 10 Graphical abstract.
Summary of interpretation of key findings from Olink measured proteins and their association with CardioResp (n = 398), Fatigue (n = 342), Anx/Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups relative to Recovered (n = 233).
Supplementary information
Supplementary information.
Supplementary Methods, Statistics and reproducibility statement, Supplementary Results, Supplementary Tables 1–7, Extended data figure legends, Appendix 1 (Supplementary Table 8), Appendix 2 (PHOSP-COVID author list) and Appendix 3 (ISARIC4C author list).
Reporting Summary
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Liew, F., Efstathiou, C., Fontanella, S. et al. Large-scale phenotyping of patients with long COVID post-hospitalization reveals mechanistic subtypes of disease. Nat Immunol 25 , 607–621 (2024). https://doi.org/10.1038/s41590-024-01778-0
Download citation
Received : 11 August 2023
Accepted : 06 February 2024
Published : 08 April 2024
Issue Date : April 2024
DOI : https://doi.org/10.1038/s41590-024-01778-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Immune dysregulation in long covid.
- Laura Ceglarek
- Onur Boyman
Nature Immunology (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors. In this Resource article, the authors integrate genomic, bioinformatic and flow cytometric data from human bone ...
Imbalanced immune responses are involved in developing preeclampsia (PE). We wish to explore the expression and potential changes of immune checkpoint molecules TIGIT, CD226 and CD155 in PE patients. Cui Li, Haiyan Liu and Zhongliang Duan. BMC Immunology 2024 25 :12. Research Published on: 7 February 2024.
Pillars of Immunology. The Journal of Immunology publishes commentaries highlighting scientific reports that were published more than 15 years ago and that have made a major impact on the course of immunological research today. Read published commentaries here . Don't miss the exciting Brief Review Collection, coming in the January 15, 2024 ...
Current Research in Immunology (CRIMMU) is an international peer-reviewed open access journal publishing original papers, short communications, and reviews that cover all aspects of molecular and cellular immunology. It is a companion to the highly regarded review journal Current Opinion in Immunology and is part of the Current Opinion and Research (CO+RE) suite of journals.
Science Immunology publishes original, peer-reviewed, science-based research articles that report critical advances in all areas of immunological research, including important new tools and techniques. mission & scope.
Immunologic Research is a transformative journal that covers the entire spectrum of immunology, from molecular and cellular levels to diseases and clinical applications. Offers both traditional publishing route and immediate gold Open Access. Maintains a balance between basic and clinical data. Provides a platform for presenting historical ...
Harmonization of ANA testing challenge: quantification strategy to accurately predict end-point titers avoiding serial dilution. Immunologic Research is a transformative journal that covers the entire spectrum of immunology, from molecular and cellular levels to diseases and clinical ...
Immunology & Cell Biology (ICB) is the flagship journal of the ASI, publishing the latest research in immunology, cellular immunology, innate and adaptive immunity, immune responses to pathogens, tumour immunology, immunopathology, immunotherapy, immunogenetics, immunological studies in humans and model organisms, and other related areas.
Immunotherapy includes the use of certain components of the immune system (antibodies, cells, cytokines, etc.) for the treatment of various cancers and autoimmune diseases and the manipulation of the immune system through vaccines for the prevention and treatment of infectious and allergic diseases (Fig. 1 ). Fig. 1.
The mammalian immune system is a remarkable sensory system for the detection and neutralization of pathogens. History is replete with the devastating effects of plagues, and the coronavirus disease 2019 (COVID-19) pandemic is a defining global health crisis of our time. Although the development of effective vaccines has saved many lives, the ...
Immunology is one of the leading journals in this field with a global representation across authors, editors and reviewers. We publish papers based on original findings in all areas of cellular and molecular immunology, and mechanistic insights into fundamental aspects of the immune system. We publish review articles on subjects of topical ...
Publications related to Immunology (10,000) Sorted by most recent Model poslovnog restrukturiranja Imunološkog zavoda d.d. iz Zagreba Business restructuring of the Immune institute from Zagreb
The rest of the paper is organized as follows. Section 4 provides a ... lymphoproliferative disorders, etc. In addition, immunology research is carried out on diseases such as cancer, mental health, bacterial infection, COVID-19, and many more. Furthermore, to understand immunology related health risks, it is essential to understand various ...
1995 ENII (European Network of Immunology Institutes) Conference Sponsored by the Europea. Read the latest articles of Research in Immunology at ScienceDirect.com, Elsevier's leading platform of peer-reviewed scholarly literature.
Invasion of spontaneous germinal centers by naive B cells is rapid and persistent. by. Theo van den Broek. Kristine Oleinika. Siti Rahmayanti. Carlos Castrillon. Cees E. van der Poel. Michael C. Carroll. Science Immunology Vol. 9, NO. 93 22 Mar 2024.
Journal of Immunology Research provides a platform for scientists and clinicians working in different and diverse areas of immunology and therapy. ... This paper reviews the importance of DC migration and current research progress in the context of DC-based immunotherapy. ... Additionally, three immune checkpoint-related genes (CD200R1, HAVCR2 ...
Cancer Immunology Research publishes outstanding original articles reporting major advances in cancer immunology that span the discipline from basic investigations in host-tumor interactions to developmental therapeutics in model systems, early translational studies in patients, and late-stage clinical trials. Read More About the Journal.
Additional authors can be found in the paper. Adapted from a Boston Children's blog post. Four new studies led by Harvard Medical School researchers at Boston Children's Hospital reveal details about how B cells in the immune system churn out antibodies that become increasingly potent and specific after we're vaccinated or exposed to an ...
Research team develops novel PTPN2/N1 inhibitor for cancer immunotherapy using generative AI. by InSilico Medicine. Insilico's compound demonstrated enhanced oral absorption, systemic exposure ...
Several research groups and companies developed chimeric and humanized mAbs (Table 2), and these mAbs included additional modifications, such as changes in the carbohydrates (glycosylation) and/or ...
BLACKSBURG, Va.--(BUSINESS WIRE)-- The NIMML Institute, ("NIMML"), a 501 (c)(3) nonprofit institute dedicated to the discovery of novel precision medicines for infectious and autoimmune diseases, today announced that it will present four abstracts regarding novel mechanistic insights related to the pharmacological activation of LANCL receptors at Immunology2024, the 107 th Annual Meeting ...
Program Director: Shiv Pillai, M.D., Ph.D., Professor of MedicineShiv Pillai is a Professor of Medicine and Health Sciences and Technology at Harvard Medical School. He is the director of the Harvard PhD and MMSc Immunology programs and of the HMS-HST MD student research program. He is also the program director of an NIH-funded Autoimmune Center of Excellence at Massachusetts General Hospital.
Abstract. Beyond structural and chemical barriers to pathogens, the immune system has two fundamental lines of defense: innate immunity and adaptive immunity. Innate immunity is the first immunological mechanism for fighting against an intruding pathogen. It is a rapid immune response, initiated within minutes or hours after aggression, that ...
Eran Ophir, Ph.D., Chief Scientific Officer at Compugen added, "Our paper published online yesterday in Cancer Immunology Research describes how COM503, a potential first-in-class high affinity ...
Introducing the Collection of Papers from the Second Workshop on Habitability of the Venus Cloud Layer and Related Research. Authors: Sanjay S. Limaye [email protected] and James B. Garvin Authors Info & Affiliations. Publication: Astrobiology. Volume 24, Issue Number 4.
Other grants that have supported this work include the UK Coronavirus Immunology Consortium (funder reference 1257927), the Imperial Biomedical Research Centre (NIHR Imperial BRC, grant IS-BRC ...