- USC Libraries
- Research Guides

Organizing Your Social Sciences Research Paper
- 6. The Methodology
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The methods section describes actions taken to investigate a research problem and the rationale for the application of specific procedures or techniques used to identify, select, process, and analyze information applied to understanding the problem, thereby, allowing the reader to critically evaluate a study’s overall validity and reliability. The methodology section of a research paper answers two main questions: How was the data collected or generated? And, how was it analyzed? The writing should be direct and precise and always written in the past tense.
Kallet, Richard H. "How to Write the Methods Section of a Research Paper." Respiratory Care 49 (October 2004): 1229-1232.
Importance of a Good Methodology Section
You must explain how you obtained and analyzed your results for the following reasons:
- Readers need to know how the data was obtained because the method you chose affects the results and, by extension, how you interpreted their significance in the discussion section of your paper.
- Methodology is crucial for any branch of scholarship because an unreliable method produces unreliable results and, as a consequence, undermines the value of your analysis of the findings.
- In most cases, there are a variety of different methods you can choose to investigate a research problem. The methodology section of your paper should clearly articulate the reasons why you have chosen a particular procedure or technique.
- The reader wants to know that the data was collected or generated in a way that is consistent with accepted practice in the field of study. For example, if you are using a multiple choice questionnaire, readers need to know that it offered your respondents a reasonable range of answers to choose from.
- The method must be appropriate to fulfilling the overall aims of the study. For example, you need to ensure that you have a large enough sample size to be able to generalize and make recommendations based upon the findings.
- The methodology should discuss the problems that were anticipated and the steps you took to prevent them from occurring. For any problems that do arise, you must describe the ways in which they were minimized or why these problems do not impact in any meaningful way your interpretation of the findings.
- In the social and behavioral sciences, it is important to always provide sufficient information to allow other researchers to adopt or replicate your methodology. This information is particularly important when a new method has been developed or an innovative use of an existing method is utilized.
Bem, Daryl J. Writing the Empirical Journal Article. Psychology Writing Center. University of Washington; Denscombe, Martyn. The Good Research Guide: For Small-Scale Social Research Projects . 5th edition. Buckingham, UK: Open University Press, 2014; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008.
Structure and Writing Style
I. Groups of Research Methods
There are two main groups of research methods in the social sciences:
- The e mpirical-analytical group approaches the study of social sciences in a similar manner that researchers study the natural sciences . This type of research focuses on objective knowledge, research questions that can be answered yes or no, and operational definitions of variables to be measured. The empirical-analytical group employs deductive reasoning that uses existing theory as a foundation for formulating hypotheses that need to be tested. This approach is focused on explanation.
- The i nterpretative group of methods is focused on understanding phenomenon in a comprehensive, holistic way . Interpretive methods focus on analytically disclosing the meaning-making practices of human subjects [the why, how, or by what means people do what they do], while showing how those practices arrange so that it can be used to generate observable outcomes. Interpretive methods allow you to recognize your connection to the phenomena under investigation. However, the interpretative group requires careful examination of variables because it focuses more on subjective knowledge.
II. Content
The introduction to your methodology section should begin by restating the research problem and underlying assumptions underpinning your study. This is followed by situating the methods you used to gather, analyze, and process information within the overall “tradition” of your field of study and within the particular research design you have chosen to study the problem. If the method you choose lies outside of the tradition of your field [i.e., your review of the literature demonstrates that the method is not commonly used], provide a justification for how your choice of methods specifically addresses the research problem in ways that have not been utilized in prior studies.
The remainder of your methodology section should describe the following:
- Decisions made in selecting the data you have analyzed or, in the case of qualitative research, the subjects and research setting you have examined,
- Tools and methods used to identify and collect information, and how you identified relevant variables,
- The ways in which you processed the data and the procedures you used to analyze that data, and
- The specific research tools or strategies that you utilized to study the underlying hypothesis and research questions.
In addition, an effectively written methodology section should:
- Introduce the overall methodological approach for investigating your research problem . Is your study qualitative or quantitative or a combination of both (mixed method)? Are you going to take a special approach, such as action research, or a more neutral stance?
- Indicate how the approach fits the overall research design . Your methods for gathering data should have a clear connection to your research problem. In other words, make sure that your methods will actually address the problem. One of the most common deficiencies found in research papers is that the proposed methodology is not suitable to achieving the stated objective of your paper.
- Describe the specific methods of data collection you are going to use , such as, surveys, interviews, questionnaires, observation, archival research. If you are analyzing existing data, such as a data set or archival documents, describe how it was originally created or gathered and by whom. Also be sure to explain how older data is still relevant to investigating the current research problem.
- Explain how you intend to analyze your results . Will you use statistical analysis? Will you use specific theoretical perspectives to help you analyze a text or explain observed behaviors? Describe how you plan to obtain an accurate assessment of relationships, patterns, trends, distributions, and possible contradictions found in the data.
- Provide background and a rationale for methodologies that are unfamiliar for your readers . Very often in the social sciences, research problems and the methods for investigating them require more explanation/rationale than widely accepted rules governing the natural and physical sciences. Be clear and concise in your explanation.
- Provide a justification for subject selection and sampling procedure . For instance, if you propose to conduct interviews, how do you intend to select the sample population? If you are analyzing texts, which texts have you chosen, and why? If you are using statistics, why is this set of data being used? If other data sources exist, explain why the data you chose is most appropriate to addressing the research problem.
- Provide a justification for case study selection . A common method of analyzing research problems in the social sciences is to analyze specific cases. These can be a person, place, event, phenomenon, or other type of subject of analysis that are either examined as a singular topic of in-depth investigation or multiple topics of investigation studied for the purpose of comparing or contrasting findings. In either method, you should explain why a case or cases were chosen and how they specifically relate to the research problem.
- Describe potential limitations . Are there any practical limitations that could affect your data collection? How will you attempt to control for potential confounding variables and errors? If your methodology may lead to problems you can anticipate, state this openly and show why pursuing this methodology outweighs the risk of these problems cropping up.
NOTE : Once you have written all of the elements of the methods section, subsequent revisions should focus on how to present those elements as clearly and as logically as possibly. The description of how you prepared to study the research problem, how you gathered the data, and the protocol for analyzing the data should be organized chronologically. For clarity, when a large amount of detail must be presented, information should be presented in sub-sections according to topic. If necessary, consider using appendices for raw data.
ANOTHER NOTE : If you are conducting a qualitative analysis of a research problem , the methodology section generally requires a more elaborate description of the methods used as well as an explanation of the processes applied to gathering and analyzing of data than is generally required for studies using quantitative methods. Because you are the primary instrument for generating the data [e.g., through interviews or observations], the process for collecting that data has a significantly greater impact on producing the findings. Therefore, qualitative research requires a more detailed description of the methods used.
YET ANOTHER NOTE : If your study involves interviews, observations, or other qualitative techniques involving human subjects , you may be required to obtain approval from the university's Office for the Protection of Research Subjects before beginning your research. This is not a common procedure for most undergraduate level student research assignments. However, i f your professor states you need approval, you must include a statement in your methods section that you received official endorsement and adequate informed consent from the office and that there was a clear assessment and minimization of risks to participants and to the university. This statement informs the reader that your study was conducted in an ethical and responsible manner. In some cases, the approval notice is included as an appendix to your paper.
III. Problems to Avoid
Irrelevant Detail The methodology section of your paper should be thorough but concise. Do not provide any background information that does not directly help the reader understand why a particular method was chosen, how the data was gathered or obtained, and how the data was analyzed in relation to the research problem [note: analyzed, not interpreted! Save how you interpreted the findings for the discussion section]. With this in mind, the page length of your methods section will generally be less than any other section of your paper except the conclusion.
Unnecessary Explanation of Basic Procedures Remember that you are not writing a how-to guide about a particular method. You should make the assumption that readers possess a basic understanding of how to investigate the research problem on their own and, therefore, you do not have to go into great detail about specific methodological procedures. The focus should be on how you applied a method , not on the mechanics of doing a method. An exception to this rule is if you select an unconventional methodological approach; if this is the case, be sure to explain why this approach was chosen and how it enhances the overall process of discovery.
Problem Blindness It is almost a given that you will encounter problems when collecting or generating your data, or, gaps will exist in existing data or archival materials. Do not ignore these problems or pretend they did not occur. Often, documenting how you overcame obstacles can form an interesting part of the methodology. It demonstrates to the reader that you can provide a cogent rationale for the decisions you made to minimize the impact of any problems that arose.
Literature Review Just as the literature review section of your paper provides an overview of sources you have examined while researching a particular topic, the methodology section should cite any sources that informed your choice and application of a particular method [i.e., the choice of a survey should include any citations to the works you used to help construct the survey].
It’s More than Sources of Information! A description of a research study's method should not be confused with a description of the sources of information. Such a list of sources is useful in and of itself, especially if it is accompanied by an explanation about the selection and use of the sources. The description of the project's methodology complements a list of sources in that it sets forth the organization and interpretation of information emanating from those sources.
Azevedo, L.F. et al. "How to Write a Scientific Paper: Writing the Methods Section." Revista Portuguesa de Pneumologia 17 (2011): 232-238; Blair Lorrie. “Choosing a Methodology.” In Writing a Graduate Thesis or Dissertation , Teaching Writing Series. (Rotterdam: Sense Publishers 2016), pp. 49-72; Butin, Dan W. The Education Dissertation A Guide for Practitioner Scholars . Thousand Oaks, CA: Corwin, 2010; Carter, Susan. Structuring Your Research Thesis . New York: Palgrave Macmillan, 2012; Kallet, Richard H. “How to Write the Methods Section of a Research Paper.” Respiratory Care 49 (October 2004):1229-1232; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008. Methods Section. The Writer’s Handbook. Writing Center. University of Wisconsin, Madison; Rudestam, Kjell Erik and Rae R. Newton. “The Method Chapter: Describing Your Research Plan.” In Surviving Your Dissertation: A Comprehensive Guide to Content and Process . (Thousand Oaks, Sage Publications, 2015), pp. 87-115; What is Interpretive Research. Institute of Public and International Affairs, University of Utah; Writing the Experimental Report: Methods, Results, and Discussion. The Writing Lab and The OWL. Purdue University; Methods and Materials. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College.
Writing Tip
Statistical Designs and Tests? Do Not Fear Them!
Don't avoid using a quantitative approach to analyzing your research problem just because you fear the idea of applying statistical designs and tests. A qualitative approach, such as conducting interviews or content analysis of archival texts, can yield exciting new insights about a research problem, but it should not be undertaken simply because you have a disdain for running a simple regression. A well designed quantitative research study can often be accomplished in very clear and direct ways, whereas, a similar study of a qualitative nature usually requires considerable time to analyze large volumes of data and a tremendous burden to create new paths for analysis where previously no path associated with your research problem had existed.
To locate data and statistics, GO HERE .
Another Writing Tip
Knowing the Relationship Between Theories and Methods
There can be multiple meaning associated with the term "theories" and the term "methods" in social sciences research. A helpful way to delineate between them is to understand "theories" as representing different ways of characterizing the social world when you research it and "methods" as representing different ways of generating and analyzing data about that social world. Framed in this way, all empirical social sciences research involves theories and methods, whether they are stated explicitly or not. However, while theories and methods are often related, it is important that, as a researcher, you deliberately separate them in order to avoid your theories playing a disproportionate role in shaping what outcomes your chosen methods produce.
Introspectively engage in an ongoing dialectic between the application of theories and methods to help enable you to use the outcomes from your methods to interrogate and develop new theories, or ways of framing conceptually the research problem. This is how scholarship grows and branches out into new intellectual territory.
Reynolds, R. Larry. Ways of Knowing. Alternative Microeconomics . Part 1, Chapter 3. Boise State University; The Theory-Method Relationship. S-Cool Revision. United Kingdom.
Yet Another Writing Tip
Methods and the Methodology
Do not confuse the terms "methods" and "methodology." As Schneider notes, a method refers to the technical steps taken to do research . Descriptions of methods usually include defining and stating why you have chosen specific techniques to investigate a research problem, followed by an outline of the procedures you used to systematically select, gather, and process the data [remember to always save the interpretation of data for the discussion section of your paper].
The methodology refers to a discussion of the underlying reasoning why particular methods were used . This discussion includes describing the theoretical concepts that inform the choice of methods to be applied, placing the choice of methods within the more general nature of academic work, and reviewing its relevance to examining the research problem. The methodology section also includes a thorough review of the methods other scholars have used to study the topic.
Bryman, Alan. "Of Methods and Methodology." Qualitative Research in Organizations and Management: An International Journal 3 (2008): 159-168; Schneider, Florian. “What's in a Methodology: The Difference between Method, Methodology, and Theory…and How to Get the Balance Right?” PoliticsEastAsia.com. Chinese Department, University of Leiden, Netherlands.
- << Previous: Scholarly vs. Popular Publications
- Next: Qualitative Methods >>
- Last Updated: May 15, 2024 9:53 AM
- URL: https://libguides.usc.edu/writingguide
Research Methodology Example
Detailed Walkthrough + Free Methodology Chapter Template
If you’re working on a dissertation or thesis and are looking for an example of a research methodology chapter , you’ve come to the right place.
In this video, we walk you through a research methodology from a dissertation that earned full distinction , step by step. We start off by discussing the core components of a research methodology by unpacking our free methodology chapter template . We then progress to the sample research methodology to show how these concepts are applied in an actual dissertation, thesis or research project.
If you’re currently working on your research methodology chapter, you may also find the following resources useful:
- Research methodology 101 : an introductory video discussing what a methodology is and the role it plays within a dissertation
- Research design 101 : an overview of the most common research designs for both qualitative and quantitative studies
- Variables 101 : an introductory video covering the different types of variables that exist within research.
- Sampling 101 : an overview of the main sampling methods
- Methodology tips : a video discussion covering various tips to help you write a high-quality methodology chapter
- Private coaching : Get hands-on help with your research methodology

PS – If you’re working on a dissertation, be sure to also check out our collection of dissertation and thesis examples here .
FAQ: Research Methodology Example
Research methodology example: frequently asked questions, is the sample research methodology real.
Yes. The chapter example is an extract from a Master’s-level dissertation for an MBA program. A few minor edits have been made to protect the privacy of the sponsoring organisation, but these have no material impact on the research methodology.
Can I replicate this methodology for my dissertation?
As we discuss in the video, every research methodology will be different, depending on the research aims, objectives and research questions. Therefore, you’ll need to tailor your literature review to suit your specific context.
You can learn more about the basics of writing a research methodology chapter here .
Where can I find more examples of research methodologies?
The best place to find more examples of methodology chapters would be within dissertation/thesis databases. These databases include dissertations, theses and research projects that have successfully passed the assessment criteria for the respective university, meaning that you have at least some sort of quality assurance.
The Open Access Thesis Database (OATD) is a good starting point.
How do I get the research methodology chapter template?
You can access our free methodology chapter template here .
Is the methodology template really free?
Yes. There is no cost for the template and you are free to use it as you wish.
You Might Also Like:

Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
- How it works
Published by Nicolas at March 21st, 2024 , Revised On March 12, 2024
The Ultimate Guide To Research Methodology
Research methodology is a crucial aspect of any investigative process, serving as the blueprint for the entire research journey. If you are stuck in the methodology section of your research paper , then this blog will guide you on what is a research methodology, its types and how to successfully conduct one.
Table of Contents
What Is Research Methodology?
Research methodology can be defined as the systematic framework that guides researchers in designing, conducting, and analyzing their investigations. It encompasses a structured set of processes, techniques, and tools employed to gather and interpret data, ensuring the reliability and validity of the research findings.
Research methodology is not confined to a singular approach; rather, it encapsulates a diverse range of methods tailored to the specific requirements of the research objectives.
Here is why Research methodology is important in academic and professional settings.
Facilitating Rigorous Inquiry
Research methodology forms the backbone of rigorous inquiry. It provides a structured approach that aids researchers in formulating precise thesis statements , selecting appropriate methodologies, and executing systematic investigations. This, in turn, enhances the quality and credibility of the research outcomes.
Ensuring Reproducibility And Reliability
In both academic and professional contexts, the ability to reproduce research outcomes is paramount. A well-defined research methodology establishes clear procedures, making it possible for others to replicate the study. This not only validates the findings but also contributes to the cumulative nature of knowledge.
Guiding Decision-Making Processes
In professional settings, decisions often hinge on reliable data and insights. Research methodology equips professionals with the tools to gather pertinent information, analyze it rigorously, and derive meaningful conclusions.
This informed decision-making is instrumental in achieving organizational goals and staying ahead in competitive environments.
Contributing To Academic Excellence
For academic researchers, adherence to robust research methodology is a hallmark of excellence. Institutions value research that adheres to high standards of methodology, fostering a culture of academic rigour and intellectual integrity. Furthermore, it prepares students with critical skills applicable beyond academia.
Enhancing Problem-Solving Abilities
Research methodology instills a problem-solving mindset by encouraging researchers to approach challenges systematically. It equips individuals with the skills to dissect complex issues, formulate hypotheses , and devise effective strategies for investigation.
Understanding Research Methodology
In the pursuit of knowledge and discovery, understanding the fundamentals of research methodology is paramount.
Basics Of Research
Research, in its essence, is a systematic and organized process of inquiry aimed at expanding our understanding of a particular subject or phenomenon. It involves the exploration of existing knowledge, the formulation of hypotheses, and the collection and analysis of data to draw meaningful conclusions.
Research is a dynamic and iterative process that contributes to the continuous evolution of knowledge in various disciplines.
Types of Research
Research takes on various forms, each tailored to the nature of the inquiry. Broadly classified, research can be categorized into two main types:
- Quantitative Research: This type involves the collection and analysis of numerical data to identify patterns, relationships, and statistical significance. It is particularly useful for testing hypotheses and making predictions.
- Qualitative Research: Qualitative research focuses on understanding the depth and details of a phenomenon through non-numerical data. It often involves methods such as interviews, focus groups, and content analysis, providing rich insights into complex issues.
Components Of Research Methodology
To conduct effective research, one must go through the different components of research methodology. These components form the scaffolding that supports the entire research process, ensuring its coherence and validity.
Research Design
Research design serves as the blueprint for the entire research project. It outlines the overall structure and strategy for conducting the study. The three primary types of research design are:
- Exploratory Research: Aimed at gaining insights and familiarity with the topic, often used in the early stages of research.
- Descriptive Research: Involves portraying an accurate profile of a situation or phenomenon, answering the ‘what,’ ‘who,’ ‘where,’ and ‘when’ questions.
- Explanatory Research: Seeks to identify the causes and effects of a phenomenon, explaining the ‘why’ and ‘how.’
Data Collection Methods
Choosing the right data collection methods is crucial for obtaining reliable and relevant information. Common methods include:
- Surveys and Questionnaires: Employed to gather information from a large number of respondents through standardized questions.
- Interviews: In-depth conversations with participants, offering qualitative insights.
- Observation: Systematic watching and recording of behaviour, events, or processes in their natural setting.
Data Analysis Techniques
Once data is collected, analysis becomes imperative to derive meaningful conclusions. Different methodologies exist for quantitative and qualitative data:
- Quantitative Data Analysis: Involves statistical techniques such as descriptive statistics, inferential statistics, and regression analysis to interpret numerical data.
- Qualitative Data Analysis: Methods like content analysis, thematic analysis, and grounded theory are employed to extract patterns, themes, and meanings from non-numerical data.
The research paper we write have:
- Precision and Clarity
- Zero Plagiarism
- High-level Encryption
- Authentic Sources
Choosing a Research Method
Selecting an appropriate research method is a critical decision in the research process. It determines the approach, tools, and techniques that will be used to answer the research questions.
Quantitative Research Methods
Quantitative research involves the collection and analysis of numerical data, providing a structured and objective approach to understanding and explaining phenomena.
Experimental Research
Experimental research involves manipulating variables to observe the effect on another variable under controlled conditions. It aims to establish cause-and-effect relationships.
Key Characteristics:
- Controlled Environment: Experiments are conducted in a controlled setting to minimize external influences.
- Random Assignment: Participants are randomly assigned to different experimental conditions.
- Quantitative Data: Data collected is numerical, allowing for statistical analysis.
Applications: Commonly used in scientific studies and psychology to test hypotheses and identify causal relationships.
Survey Research
Survey research gathers information from a sample of individuals through standardized questionnaires or interviews. It aims to collect data on opinions, attitudes, and behaviours.
- Structured Instruments: Surveys use structured instruments, such as questionnaires, to collect data.
- Large Sample Size: Surveys often target a large and diverse group of participants.
- Quantitative Data Analysis: Responses are quantified for statistical analysis.
Applications: Widely employed in social sciences, marketing, and public opinion research to understand trends and preferences.
Descriptive Research
Descriptive research seeks to portray an accurate profile of a situation or phenomenon. It focuses on answering the ‘what,’ ‘who,’ ‘where,’ and ‘when’ questions.
- Observation and Data Collection: This involves observing and documenting without manipulating variables.
- Objective Description: Aim to provide an unbiased and factual account of the subject.
- Quantitative or Qualitative Data: T his can include both types of data, depending on the research focus.
Applications: Useful in situations where researchers want to understand and describe a phenomenon without altering it, common in social sciences and education.
Qualitative Research Methods
Qualitative research emphasizes exploring and understanding the depth and complexity of phenomena through non-numerical data.
A case study is an in-depth exploration of a particular person, group, event, or situation. It involves detailed, context-rich analysis.
- Rich Data Collection: Uses various data sources, such as interviews, observations, and documents.
- Contextual Understanding: Aims to understand the context and unique characteristics of the case.
- Holistic Approach: Examines the case in its entirety.
Applications: Common in social sciences, psychology, and business to investigate complex and specific instances.
Ethnography
Ethnography involves immersing the researcher in the culture or community being studied to gain a deep understanding of their behaviours, beliefs, and practices.
- Participant Observation: Researchers actively participate in the community or setting.
- Holistic Perspective: Focuses on the interconnectedness of cultural elements.
- Qualitative Data: In-depth narratives and descriptions are central to ethnographic studies.
Applications: Widely used in anthropology, sociology, and cultural studies to explore and document cultural practices.
Grounded Theory
Grounded theory aims to develop theories grounded in the data itself. It involves systematic data collection and analysis to construct theories from the ground up.
- Constant Comparison: Data is continually compared and analyzed during the research process.
- Inductive Reasoning: Theories emerge from the data rather than being imposed on it.
- Iterative Process: The research design evolves as the study progresses.
Applications: Commonly applied in sociology, nursing, and management studies to generate theories from empirical data.
Research design is the structural framework that outlines the systematic process and plan for conducting a study. It serves as the blueprint, guiding researchers on how to collect, analyze, and interpret data.
Exploratory, Descriptive, And Explanatory Designs
Exploratory design.
Exploratory research design is employed when a researcher aims to explore a relatively unknown subject or gain insights into a complex phenomenon.
- Flexibility: Allows for flexibility in data collection and analysis.
- Open-Ended Questions: Uses open-ended questions to gather a broad range of information.
- Preliminary Nature: Often used in the initial stages of research to formulate hypotheses.
Applications: Valuable in the early stages of investigation, especially when the researcher seeks a deeper understanding of a subject before formalizing research questions.
Descriptive Design
Descriptive research design focuses on portraying an accurate profile of a situation, group, or phenomenon.
- Structured Data Collection: Involves systematic and structured data collection methods.
- Objective Presentation: Aims to provide an unbiased and factual account of the subject.
- Quantitative or Qualitative Data: Can incorporate both types of data, depending on the research objectives.
Applications: Widely used in social sciences, marketing, and educational research to provide detailed and objective descriptions.
Explanatory Design
Explanatory research design aims to identify the causes and effects of a phenomenon, explaining the ‘why’ and ‘how’ behind observed relationships.
- Causal Relationships: Seeks to establish causal relationships between variables.
- Controlled Variables : Often involves controlling certain variables to isolate causal factors.
- Quantitative Analysis: Primarily relies on quantitative data analysis techniques.
Applications: Commonly employed in scientific studies and social sciences to delve into the underlying reasons behind observed patterns.
Cross-Sectional Vs. Longitudinal Designs
Cross-sectional design.
Cross-sectional designs collect data from participants at a single point in time.
- Snapshot View: Provides a snapshot of a population at a specific moment.
- Efficiency: More efficient in terms of time and resources.
- Limited Temporal Insights: Offers limited insights into changes over time.
Applications: Suitable for studying characteristics or behaviours that are stable or not expected to change rapidly.
Longitudinal Design
Longitudinal designs involve the collection of data from the same participants over an extended period.
- Temporal Sequence: Allows for the examination of changes over time.
- Causality Assessment: Facilitates the assessment of cause-and-effect relationships.
- Resource-Intensive: Requires more time and resources compared to cross-sectional designs.
Applications: Ideal for studying developmental processes, trends, or the impact of interventions over time.
Experimental Vs Non-experimental Designs
Experimental design.
Experimental designs involve manipulating variables under controlled conditions to observe the effect on another variable.
- Causality Inference: Enables the inference of cause-and-effect relationships.
- Quantitative Data: Primarily involves the collection and analysis of numerical data.
Applications: Commonly used in scientific studies, psychology, and medical research to establish causal relationships.
Non-Experimental Design
Non-experimental designs observe and describe phenomena without manipulating variables.
- Natural Settings: Data is often collected in natural settings without intervention.
- Descriptive or Correlational: Focuses on describing relationships or correlations between variables.
- Quantitative or Qualitative Data: This can involve either type of data, depending on the research approach.
Applications: Suitable for studying complex phenomena in real-world settings where manipulation may not be ethical or feasible.
Effective data collection is fundamental to the success of any research endeavour.
Designing Effective Surveys
Objective Design:
- Clearly define the research objectives to guide the survey design.
- Craft questions that align with the study’s goals and avoid ambiguity.
Structured Format:
- Use a structured format with standardized questions for consistency.
- Include a mix of closed-ended and open-ended questions for detailed insights.
Pilot Testing:
- Conduct pilot tests to identify and rectify potential issues with survey design.
- Ensure clarity, relevance, and appropriateness of questions.
Sampling Strategy:
- Develop a robust sampling strategy to ensure a representative participant group.
- Consider random sampling or stratified sampling based on the research goals.
Conducting Interviews
Establishing Rapport:
- Build rapport with participants to create a comfortable and open environment.
- Clearly communicate the purpose of the interview and the value of participants’ input.
Open-Ended Questions:
- Frame open-ended questions to encourage detailed responses.
- Allow participants to express their thoughts and perspectives freely.
Active Listening:
- Practice active listening to understand areas and gather rich data.
- Avoid interrupting and maintain a non-judgmental stance during the interview.
Ethical Considerations:
- Obtain informed consent and assure participants of confidentiality.
- Be transparent about the study’s purpose and potential implications.
Observation
1. participant observation.
Immersive Participation:
- Actively immerse yourself in the setting or group being observed.
- Develop a deep understanding of behaviours, interactions, and context.
Field Notes:
- Maintain detailed and reflective field notes during observations.
- Document observed patterns, unexpected events, and participant reactions.
Ethical Awareness:
- Be conscious of ethical considerations, ensuring respect for participants.
- Balance the role of observer and participant to minimize bias.
2. Non-participant Observation
Objective Observation:
- Maintain a more detached and objective stance during non-participant observation.
- Focus on recording behaviours, events, and patterns without direct involvement.
Data Reliability:
- Enhance the reliability of data by reducing observer bias.
- Develop clear observation protocols and guidelines.
Contextual Understanding:
- Strive for a thorough understanding of the observed context.
- Consider combining non-participant observation with other methods for triangulation.
Archival Research
1. using existing data.
Identifying Relevant Archives:
- Locate and access archives relevant to the research topic.
- Collaborate with institutions or repositories holding valuable data.
Data Verification:
- Verify the accuracy and reliability of archived data.
- Cross-reference with other sources to ensure data integrity.
Ethical Use:
- Adhere to ethical guidelines when using existing data.
- Respect copyright and intellectual property rights.
2. Challenges and Considerations
Incomplete or Inaccurate Archives:
- Address the possibility of incomplete or inaccurate archival records.
- Acknowledge limitations and uncertainties in the data.
Temporal Bias:
- Recognize potential temporal biases in archived data.
- Consider the historical context and changes that may impact interpretation.
Access Limitations:
- Address potential limitations in accessing certain archives.
- Seek alternative sources or collaborate with institutions to overcome barriers.
Common Challenges in Research Methodology
Conducting research is a complex and dynamic process, often accompanied by a myriad of challenges. Addressing these challenges is crucial to ensure the reliability and validity of research findings.
Sampling Issues
Sampling bias:.
- The presence of sampling bias can lead to an unrepresentative sample, affecting the generalizability of findings.
- Employ random sampling methods and ensure the inclusion of diverse participants to reduce bias.
Sample Size Determination:
- Determining an appropriate sample size is a delicate balance. Too small a sample may lack statistical power, while an excessively large sample may strain resources.
- Conduct a power analysis to determine the optimal sample size based on the research objectives and expected effect size.
Data Quality And Validity
Measurement error:.
- Inaccuracies in measurement tools or data collection methods can introduce measurement errors, impacting the validity of results.
- Pilot test instruments, calibrate equipment, and use standardized measures to enhance the reliability of data.
Construct Validity:
- Ensuring that the chosen measures accurately capture the intended constructs is a persistent challenge.
- Use established measurement instruments and employ multiple measures to assess the same construct for triangulation.
Time And Resource Constraints
Timeline pressures:.
- Limited timeframes can compromise the depth and thoroughness of the research process.
- Develop a realistic timeline, prioritize tasks, and communicate expectations with stakeholders to manage time constraints effectively.
Resource Availability:
- Inadequate resources, whether financial or human, can impede the execution of research activities.
- Seek external funding, collaborate with other researchers, and explore alternative methods that require fewer resources.
Managing Bias in Research
Selection bias:.
- Selecting participants in a way that systematically skews the sample can introduce selection bias.
- Employ randomization techniques, use stratified sampling, and transparently report participant recruitment methods.
Confirmation Bias:
- Researchers may unintentionally favour information that confirms their preconceived beliefs or hypotheses.
- Adopt a systematic and open-minded approach, use blinded study designs, and engage in peer review to mitigate confirmation bias.
Tips On How To Write A Research Methodology
Conducting successful research relies not only on the application of sound methodologies but also on strategic planning and effective collaboration. Here are some tips to enhance the success of your research methodology:
Tip 1. Clear Research Objectives
Well-defined research objectives guide the entire research process. Clearly articulate the purpose of your study, outlining specific research questions or hypotheses.
Tip 2. Comprehensive Literature Review
A thorough literature review provides a foundation for understanding existing knowledge and identifying gaps. Invest time in reviewing relevant literature to inform your research design and methodology.
Tip 3. Detailed Research Plan
A detailed plan serves as a roadmap, ensuring all aspects of the research are systematically addressed. Develop a detailed research plan outlining timelines, milestones, and tasks.
Tip 4. Ethical Considerations
Ethical practices are fundamental to maintaining the integrity of research. Address ethical considerations early, obtain necessary approvals, and ensure participant rights are safeguarded.
Tip 5. Stay Updated On Methodologies
Research methodologies evolve, and staying updated is essential for employing the most effective techniques. Engage in continuous learning by attending workshops, conferences, and reading recent publications.
Tip 6. Adaptability In Methods
Unforeseen challenges may arise during research, necessitating adaptability in methods. Be flexible and willing to modify your approach when needed, ensuring the integrity of the study.
Tip 7. Iterative Approach
Research is often an iterative process, and refining methods based on ongoing findings enhance the study’s robustness. Regularly review and refine your research design and methods as the study progresses.
Frequently Asked Questions
What is the research methodology.
Research methodology is the systematic process of planning, executing, and evaluating scientific investigation. It encompasses the techniques, tools, and procedures used to collect, analyze, and interpret data, ensuring the reliability and validity of research findings.
What are the methodologies in research?
Research methodologies include qualitative and quantitative approaches. Qualitative methods involve in-depth exploration of non-numerical data, while quantitative methods use statistical analysis to examine numerical data. Mixed methods combine both approaches for a comprehensive understanding of research questions.
How to write research methodology?
To write a research methodology, clearly outline the study’s design, data collection, and analysis procedures. Specify research tools, participants, and sampling methods. Justify choices and discuss limitations. Ensure clarity, coherence, and alignment with research objectives for a robust methodology section.
How to write the methodology section of a research paper?
In the methodology section of a research paper, describe the study’s design, data collection, and analysis methods. Detail procedures, tools, participants, and sampling. Justify choices, address ethical considerations, and explain how the methodology aligns with research objectives, ensuring clarity and rigour.
What is mixed research methodology?
Mixed research methodology combines both qualitative and quantitative research approaches within a single study. This approach aims to enhance the details and depth of research findings by providing a more comprehensive understanding of the research problem or question.
You May Also Like
The central idea of this excerpt revolves around the exploration of key themes, offering insights that illuminate the concepts within the text.
Common topics in Botany papers include taxonomy, plant physiology, ecology and biodiversity, plant pathology, and genetics.
Craft a compelling scholarship motivation letter by showcasing your passion, achievements, and future goals concisely and impactfully.
Ready to place an order?
USEFUL LINKS
Learning resources, company details.
- How It Works
Automated page speed optimizations for fast site performance
Get science-backed answers as you write with Paperpal's Research feature
What is Research Methodology? Definition, Types, and Examples

Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of the research. Several aspects must be considered before selecting an appropriate research methodology, such as research limitations and ethical concerns that may affect your research.
The research methodology section in a scientific paper describes the different methodological choices made, such as the data collection and analysis methods, and why these choices were selected. The reasons should explain why the methods chosen are the most appropriate to answer the research question. A good research methodology also helps ensure the reliability and validity of the research findings. There are three types of research methodology—quantitative, qualitative, and mixed-method, which can be chosen based on the research objectives.
What is research methodology ?
A research methodology describes the techniques and procedures used to identify and analyze information regarding a specific research topic. It is a process by which researchers design their study so that they can achieve their objectives using the selected research instruments. It includes all the important aspects of research, including research design, data collection methods, data analysis methods, and the overall framework within which the research is conducted. While these points can help you understand what is research methodology, you also need to know why it is important to pick the right methodology.
Why is research methodology important?
Having a good research methodology in place has the following advantages: 3
- Helps other researchers who may want to replicate your research; the explanations will be of benefit to them.
- You can easily answer any questions about your research if they arise at a later stage.
- A research methodology provides a framework and guidelines for researchers to clearly define research questions, hypotheses, and objectives.
- It helps researchers identify the most appropriate research design, sampling technique, and data collection and analysis methods.
- A sound research methodology helps researchers ensure that their findings are valid and reliable and free from biases and errors.
- It also helps ensure that ethical guidelines are followed while conducting research.
- A good research methodology helps researchers in planning their research efficiently, by ensuring optimum usage of their time and resources.
Writing the methods section of a research paper? Let Paperpal help you achieve perfection
Types of research methodology.
There are three types of research methodology based on the type of research and the data required. 1
- Quantitative research methodology focuses on measuring and testing numerical data. This approach is good for reaching a large number of people in a short amount of time. This type of research helps in testing the causal relationships between variables, making predictions, and generalizing results to wider populations.
- Qualitative research methodology examines the opinions, behaviors, and experiences of people. It collects and analyzes words and textual data. This research methodology requires fewer participants but is still more time consuming because the time spent per participant is quite large. This method is used in exploratory research where the research problem being investigated is not clearly defined.
- Mixed-method research methodology uses the characteristics of both quantitative and qualitative research methodologies in the same study. This method allows researchers to validate their findings, verify if the results observed using both methods are complementary, and explain any unexpected results obtained from one method by using the other method.
What are the types of sampling designs in research methodology?
Sampling 4 is an important part of a research methodology and involves selecting a representative sample of the population to conduct the study, making statistical inferences about them, and estimating the characteristics of the whole population based on these inferences. There are two types of sampling designs in research methodology—probability and nonprobability.
- Probability sampling
In this type of sampling design, a sample is chosen from a larger population using some form of random selection, that is, every member of the population has an equal chance of being selected. The different types of probability sampling are:
- Systematic —sample members are chosen at regular intervals. It requires selecting a starting point for the sample and sample size determination that can be repeated at regular intervals. This type of sampling method has a predefined range; hence, it is the least time consuming.
- Stratified —researchers divide the population into smaller groups that don’t overlap but represent the entire population. While sampling, these groups can be organized, and then a sample can be drawn from each group separately.
- Cluster —the population is divided into clusters based on demographic parameters like age, sex, location, etc.
- Convenience —selects participants who are most easily accessible to researchers due to geographical proximity, availability at a particular time, etc.
- Purposive —participants are selected at the researcher’s discretion. Researchers consider the purpose of the study and the understanding of the target audience.
- Snowball —already selected participants use their social networks to refer the researcher to other potential participants.
- Quota —while designing the study, the researchers decide how many people with which characteristics to include as participants. The characteristics help in choosing people most likely to provide insights into the subject.
What are data collection methods?
During research, data are collected using various methods depending on the research methodology being followed and the research methods being undertaken. Both qualitative and quantitative research have different data collection methods, as listed below.
Qualitative research 5
- One-on-one interviews: Helps the interviewers understand a respondent’s subjective opinion and experience pertaining to a specific topic or event
- Document study/literature review/record keeping: Researchers’ review of already existing written materials such as archives, annual reports, research articles, guidelines, policy documents, etc.
- Focus groups: Constructive discussions that usually include a small sample of about 6-10 people and a moderator, to understand the participants’ opinion on a given topic.
- Qualitative observation : Researchers collect data using their five senses (sight, smell, touch, taste, and hearing).
Quantitative research 6
- Sampling: The most common type is probability sampling.
- Interviews: Commonly telephonic or done in-person.
- Observations: Structured observations are most commonly used in quantitative research. In this method, researchers make observations about specific behaviors of individuals in a structured setting.
- Document review: Reviewing existing research or documents to collect evidence for supporting the research.
- Surveys and questionnaires. Surveys can be administered both online and offline depending on the requirement and sample size.
Let Paperpal help you write the perfect research methods section. Start now!
What are data analysis methods.
The data collected using the various methods for qualitative and quantitative research need to be analyzed to generate meaningful conclusions. These data analysis methods 7 also differ between quantitative and qualitative research.
Quantitative research involves a deductive method for data analysis where hypotheses are developed at the beginning of the research and precise measurement is required. The methods include statistical analysis applications to analyze numerical data and are grouped into two categories—descriptive and inferential.
Descriptive analysis is used to describe the basic features of different types of data to present it in a way that ensures the patterns become meaningful. The different types of descriptive analysis methods are:
- Measures of frequency (count, percent, frequency)
- Measures of central tendency (mean, median, mode)
- Measures of dispersion or variation (range, variance, standard deviation)
- Measure of position (percentile ranks, quartile ranks)
Inferential analysis is used to make predictions about a larger population based on the analysis of the data collected from a smaller population. This analysis is used to study the relationships between different variables. Some commonly used inferential data analysis methods are:
- Correlation: To understand the relationship between two or more variables.
- Cross-tabulation: Analyze the relationship between multiple variables.
- Regression analysis: Study the impact of independent variables on the dependent variable.
- Frequency tables: To understand the frequency of data.
- Analysis of variance: To test the degree to which two or more variables differ in an experiment.
Qualitative research involves an inductive method for data analysis where hypotheses are developed after data collection. The methods include:
- Content analysis: For analyzing documented information from text and images by determining the presence of certain words or concepts in texts.
- Narrative analysis: For analyzing content obtained from sources such as interviews, field observations, and surveys. The stories and opinions shared by people are used to answer research questions.
- Discourse analysis: For analyzing interactions with people considering the social context, that is, the lifestyle and environment, under which the interaction occurs.
- Grounded theory: Involves hypothesis creation by data collection and analysis to explain why a phenomenon occurred.
- Thematic analysis: To identify important themes or patterns in data and use these to address an issue.
How to choose a research methodology?
Here are some important factors to consider when choosing a research methodology: 8
- Research objectives, aims, and questions —these would help structure the research design.
- Review existing literature to identify any gaps in knowledge.
- Check the statistical requirements —if data-driven or statistical results are needed then quantitative research is the best. If the research questions can be answered based on people’s opinions and perceptions, then qualitative research is most suitable.
- Sample size —sample size can often determine the feasibility of a research methodology. For a large sample, less effort- and time-intensive methods are appropriate.
- Constraints —constraints of time, geography, and resources can help define the appropriate methodology.
Got writer’s block? Kickstart your research paper writing with Paperpal now!
How to write a research methodology .
A research methodology should include the following components: 3,9
- Research design —should be selected based on the research question and the data required. Common research designs include experimental, quasi-experimental, correlational, descriptive, and exploratory.
- Research method —this can be quantitative, qualitative, or mixed-method.
- Reason for selecting a specific methodology —explain why this methodology is the most suitable to answer your research problem.
- Research instruments —explain the research instruments you plan to use, mainly referring to the data collection methods such as interviews, surveys, etc. Here as well, a reason should be mentioned for selecting the particular instrument.
- Sampling —this involves selecting a representative subset of the population being studied.
- Data collection —involves gathering data using several data collection methods, such as surveys, interviews, etc.
- Data analysis —describe the data analysis methods you will use once you’ve collected the data.
- Research limitations —mention any limitations you foresee while conducting your research.
- Validity and reliability —validity helps identify the accuracy and truthfulness of the findings; reliability refers to the consistency and stability of the results over time and across different conditions.
- Ethical considerations —research should be conducted ethically. The considerations include obtaining consent from participants, maintaining confidentiality, and addressing conflicts of interest.
Streamline Your Research Paper Writing Process with Paperpal
The methods section is a critical part of the research papers, allowing researchers to use this to understand your findings and replicate your work when pursuing their own research. However, it is usually also the most difficult section to write. This is where Paperpal can help you overcome the writer’s block and create the first draft in minutes with Paperpal Copilot, its secure generative AI feature suite.
With Paperpal you can get research advice, write and refine your work, rephrase and verify the writing, and ensure submission readiness, all in one place. Here’s how you can use Paperpal to develop the first draft of your methods section.
- Generate an outline: Input some details about your research to instantly generate an outline for your methods section
- Develop the section: Use the outline and suggested sentence templates to expand your ideas and develop the first draft.
- P araph ras e and trim : Get clear, concise academic text with paraphrasing that conveys your work effectively and word reduction to fix redundancies.
- Choose the right words: Enhance text by choosing contextual synonyms based on how the words have been used in previously published work.
- Check and verify text : Make sure the generated text showcases your methods correctly, has all the right citations, and is original and authentic. .
You can repeat this process to develop each section of your research manuscript, including the title, abstract and keywords. Ready to write your research papers faster, better, and without the stress? Sign up for Paperpal and start writing today!
Frequently Asked Questions
Q1. What are the key components of research methodology?
A1. A good research methodology has the following key components:
- Research design
- Data collection procedures
- Data analysis methods
- Ethical considerations
Q2. Why is ethical consideration important in research methodology?
A2. Ethical consideration is important in research methodology to ensure the readers of the reliability and validity of the study. Researchers must clearly mention the ethical norms and standards followed during the conduct of the research and also mention if the research has been cleared by any institutional board. The following 10 points are the important principles related to ethical considerations: 10
- Participants should not be subjected to harm.
- Respect for the dignity of participants should be prioritized.
- Full consent should be obtained from participants before the study.
- Participants’ privacy should be ensured.
- Confidentiality of the research data should be ensured.
- Anonymity of individuals and organizations participating in the research should be maintained.
- The aims and objectives of the research should not be exaggerated.
- Affiliations, sources of funding, and any possible conflicts of interest should be declared.
- Communication in relation to the research should be honest and transparent.
- Misleading information and biased representation of primary data findings should be avoided.
Q3. What is the difference between methodology and method?
A3. Research methodology is different from a research method, although both terms are often confused. Research methods are the tools used to gather data, while the research methodology provides a framework for how research is planned, conducted, and analyzed. The latter guides researchers in making decisions about the most appropriate methods for their research. Research methods refer to the specific techniques, procedures, and tools used by researchers to collect, analyze, and interpret data, for instance surveys, questionnaires, interviews, etc.
Research methodology is, thus, an integral part of a research study. It helps ensure that you stay on track to meet your research objectives and answer your research questions using the most appropriate data collection and analysis tools based on your research design.
Accelerate your research paper writing with Paperpal. Try for free now!
- Research methodologies. Pfeiffer Library website. Accessed August 15, 2023. https://library.tiffin.edu/researchmethodologies/whatareresearchmethodologies
- Types of research methodology. Eduvoice website. Accessed August 16, 2023. https://eduvoice.in/types-research-methodology/
- The basics of research methodology: A key to quality research. Voxco. Accessed August 16, 2023. https://www.voxco.com/blog/what-is-research-methodology/
- Sampling methods: Types with examples. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/types-of-sampling-for-social-research/
- What is qualitative research? Methods, types, approaches, examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-qualitative-research-methods-types-examples/
- What is quantitative research? Definition, methods, types, and examples. Researcher.Life blog. Accessed August 15, 2023. https://researcher.life/blog/article/what-is-quantitative-research-types-and-examples/
- Data analysis in research: Types & methods. QuestionPro website. Accessed August 16, 2023. https://www.questionpro.com/blog/data-analysis-in-research/#Data_analysis_in_qualitative_research
- Factors to consider while choosing the right research methodology. PhD Monster website. Accessed August 17, 2023. https://www.phdmonster.com/factors-to-consider-while-choosing-the-right-research-methodology/
- What is research methodology? Research and writing guides. Accessed August 14, 2023. https://paperpile.com/g/what-is-research-methodology/
- Ethical considerations. Business research methodology website. Accessed August 17, 2023. https://research-methodology.net/research-methodology/ethical-considerations/
Paperpal is a comprehensive AI writing toolkit that helps students and researchers achieve 2x the writing in half the time. It leverages 21+ years of STM experience and insights from millions of research articles to provide in-depth academic writing, language editing, and submission readiness support to help you write better, faster.
Get accurate academic translations, rewriting support, grammar checks, vocabulary suggestions, and generative AI assistance that delivers human precision at machine speed. Try for free or upgrade to Paperpal Prime starting at US$19 a month to access premium features, including consistency, plagiarism, and 30+ submission readiness checks to help you succeed.
Experience the future of academic writing – Sign up to Paperpal and start writing for free!
Related Reads:
- Dangling Modifiers and How to Avoid Them in Your Writing
- Webinar: How to Use Generative AI Tools Ethically in Your Academic Writing
- Research Outlines: How to Write An Introduction Section in Minutes with Paperpal Copilot
- How to Paraphrase Research Papers Effectively
Language and Grammar Rules for Academic Writing
Climatic vs. climactic: difference and examples, you may also like, how to write a high-quality conference paper, how paperpal is enhancing academic productivity and accelerating..., academic editing: how to self-edit academic text with..., 4 ways paperpal encourages responsible writing with ai, what are scholarly sources and where can you..., how to write a hypothesis types and examples , what is academic writing: tips for students, what is hedging in academic writing , how to use ai to enhance your college..., how to use paperpal to generate emails &....
- PRO Courses Guides New Tech Help Pro Expert Videos About wikiHow Pro Upgrade Sign In
- EDIT Edit this Article
- EXPLORE Tech Help Pro About Us Random Article Quizzes Request a New Article Community Dashboard This Or That Game Popular Categories Arts and Entertainment Artwork Books Movies Computers and Electronics Computers Phone Skills Technology Hacks Health Men's Health Mental Health Women's Health Relationships Dating Love Relationship Issues Hobbies and Crafts Crafts Drawing Games Education & Communication Communication Skills Personal Development Studying Personal Care and Style Fashion Hair Care Personal Hygiene Youth Personal Care School Stuff Dating All Categories Arts and Entertainment Finance and Business Home and Garden Relationship Quizzes Cars & Other Vehicles Food and Entertaining Personal Care and Style Sports and Fitness Computers and Electronics Health Pets and Animals Travel Education & Communication Hobbies and Crafts Philosophy and Religion Work World Family Life Holidays and Traditions Relationships Youth
- Browse Articles
- Learn Something New
- Quizzes Hot
- This Or That Game
- Train Your Brain
- Explore More
- Support wikiHow
- About wikiHow
- Log in / Sign up
- Education and Communications
- College University and Postgraduate
- Academic Writing
How to Write Research Methodology
Last Updated: May 21, 2023 Approved
This article was co-authored by Alexander Ruiz, M.Ed. and by wikiHow staff writer, Jennifer Mueller, JD . Alexander Ruiz is an Educational Consultant and the Educational Director of Link Educational Institute, a tutoring business based in Claremont, California that provides customizable educational plans, subject and test prep tutoring, and college application consulting. With over a decade and a half of experience in the education industry, Alexander coaches students to increase their self-awareness and emotional intelligence while achieving skills and the goal of achieving skills and higher education. He holds a BA in Psychology from Florida International University and an MA in Education from Georgia Southern University. wikiHow marks an article as reader-approved once it receives enough positive feedback. In this case, several readers have written to tell us that this article was helpful to them, earning it our reader-approved status. This article has been viewed 521,567 times.
The research methodology section of any academic research paper gives you the opportunity to convince your readers that your research is useful and will contribute to your field of study. An effective research methodology is grounded in your overall approach – whether qualitative or quantitative – and adequately describes the methods you used. Justify why you chose those methods over others, then explain how those methods will provide answers to your research questions. [1] X Research source
Describing Your Methods

- In your restatement, include any underlying assumptions that you're making or conditions that you're taking for granted. These assumptions will also inform the research methods you've chosen.
- Generally, state the variables you'll test and the other conditions you're controlling or assuming are equal.

- If you want to research and document measurable social trends, or evaluate the impact of a particular policy on various variables, use a quantitative approach focused on data collection and statistical analysis.
- If you want to evaluate people's views or understanding of a particular issue, choose a more qualitative approach.
- You can also combine the two. For example, you might look primarily at a measurable social trend, but also interview people and get their opinions on how that trend is affecting their lives.

- For example, if you conducted a survey, you would describe the questions included in the survey, where and how the survey was conducted (such as in person, online, over the phone), how many surveys were distributed, and how long your respondents had to complete the survey.
- Include enough detail that your study can be replicated by others in your field, even if they may not get the same results you did. [4] X Research source

- Qualitative research methods typically require more detailed explanation than quantitative methods.
- Basic investigative procedures don't need to be explained in detail. Generally, you can assume that your readers have a general understanding of common research methods that social scientists use, such as surveys or focus groups.

- For example, suppose you conducted a survey and used a couple of other research papers to help construct the questions on your survey. You would mention those as contributing sources.
Justifying Your Choice of Methods

- Describe study participants specifically, and list any inclusion or exclusion criteria you used when forming your group of participants.
- Justify the size of your sample, if applicable, and describe how this affects whether your study can be generalized to larger populations. For example, if you conducted a survey of 30 percent of the student population of a university, you could potentially apply those results to the student body as a whole, but maybe not to students at other universities.

- Reading other research papers is a good way to identify potential problems that commonly arise with various methods. State whether you actually encountered any of these common problems during your research.

- If you encountered any problems as you collected data, explain clearly the steps you took to minimize the effect that problem would have on your results.

- In some cases, this may be as simple as stating that while there were numerous studies using one method, there weren't any using your method, which caused a gap in understanding of the issue.
- For example, there may be multiple papers providing quantitative analysis of a particular social trend. However, none of these papers looked closely at how this trend was affecting the lives of people.
Connecting Your Methods to Your Research Goals

- Depending on your research questions, you may be mixing quantitative and qualitative analysis – just as you could potentially use both approaches. For example, you might do a statistical analysis, and then interpret those statistics through a particular theoretical lens.

- For example, suppose you're researching the effect of college education on family farms in rural America. While you could do interviews of college-educated people who grew up on a family farm, that would not give you a picture of the overall effect. A quantitative approach and statistical analysis would give you a bigger picture.

- If in answering your research questions, your findings have raised other questions that may require further research, state these briefly.
- You can also include here any limitations to your methods, or questions that weren't answered through your research.

- Generalization is more typically used in quantitative research. If you have a well-designed sample, you can statistically apply your results to the larger population your sample belongs to.
Template to Write Research Methodology

Community Q&A
- Organize your methodology section chronologically, starting with how you prepared to conduct your research methods, how you gathered data, and how you analyzed that data. [13] X Research source Thanks Helpful 0 Not Helpful 0
- Write your research methodology section in past tense, unless you're submitting the methodology section before the research described has been carried out. [14] X Research source Thanks Helpful 2 Not Helpful 0
- Discuss your plans in detail with your advisor or supervisor before committing to a particular methodology. They can help identify possible flaws in your study. [15] X Research source Thanks Helpful 0 Not Helpful 0

You Might Also Like

- ↑ http://expertjournals.com/how-to-write-a-research-methodology-for-your-academic-article/
- ↑ http://libguides.usc.edu/writingguide/methodology
- ↑ https://www.skillsyouneed.com/learn/dissertation-methodology.html
- ↑ https://uir.unisa.ac.za/bitstream/handle/10500/4245/05Chap%204_Research%20methodology%20and%20design.pdf
- ↑ https://elc.polyu.edu.hk/FYP/html/method.htm
About This Article

To write a research methodology, start with a section that outlines the problems or questions you'll be studying, including your hypotheses or whatever it is you're setting out to prove. Then, briefly explain why you chose to use either a qualitative or quantitative approach for your study. Next, go over when and where you conducted your research and what parameters you used to ensure you were objective. Finally, cite any sources you used to decide on the methodology for your research. To learn how to justify your choice of methods in your research methodology, scroll down! Did this summary help you? Yes No
- Send fan mail to authors
Reader Success Stories
Prof. Dr. Ahmed Askar
Apr 18, 2020
Did this article help you?

M. Mahmood Shah Khan
Mar 17, 2020
Shimola Makondo
Jul 20, 2019
Zain Sharif Mohammed Alnadhery
Jan 7, 2019
Lundi Dukashe
Feb 17, 2020

Featured Articles

Trending Articles

Watch Articles

- Terms of Use
- Privacy Policy
- Do Not Sell or Share My Info
- Not Selling Info
Get all the best how-tos!
Sign up for wikiHow's weekly email newsletter
- Privacy Policy

Home » How To Write A Proposal – Step By Step Guide [With Template]
How To Write A Proposal – Step By Step Guide [With Template]
Table of Contents

How To Write A Proposal
Writing a Proposal involves several key steps to effectively communicate your ideas and intentions to a target audience. Here’s a detailed breakdown of each step:
Identify the Purpose and Audience
- Clearly define the purpose of your proposal: What problem are you addressing, what solution are you proposing, or what goal are you aiming to achieve?
- Identify your target audience: Who will be reading your proposal? Consider their background, interests, and any specific requirements they may have.
Conduct Research
- Gather relevant information: Conduct thorough research to support your proposal. This may involve studying existing literature, analyzing data, or conducting surveys/interviews to gather necessary facts and evidence.
- Understand the context: Familiarize yourself with the current situation or problem you’re addressing. Identify any relevant trends, challenges, or opportunities that may impact your proposal.
Develop an Outline
- Create a clear and logical structure: Divide your proposal into sections or headings that will guide your readers through the content.
- Introduction: Provide a concise overview of the problem, its significance, and the proposed solution.
- Background/Context: Offer relevant background information and context to help the readers understand the situation.
- Objectives/Goals: Clearly state the objectives or goals of your proposal.
- Methodology/Approach: Describe the approach or methodology you will use to address the problem.
- Timeline/Schedule: Present a detailed timeline or schedule outlining the key milestones or activities.
- Budget/Resources: Specify the financial and other resources required to implement your proposal.
- Evaluation/Success Metrics: Explain how you will measure the success or effectiveness of your proposal.
- Conclusion: Summarize the main points and restate the benefits of your proposal.
Write the Proposal
- Grab attention: Start with a compelling opening statement or a brief story that hooks the reader.
- Clearly state the problem: Clearly define the problem or issue you are addressing and explain its significance.
- Present your proposal: Introduce your proposed solution, project, or idea and explain why it is the best approach.
- State the objectives/goals: Clearly articulate the specific objectives or goals your proposal aims to achieve.
- Provide supporting information: Present evidence, data, or examples to support your claims and justify your proposal.
- Explain the methodology: Describe in detail the approach, methods, or strategies you will use to implement your proposal.
- Address potential concerns: Anticipate and address any potential objections or challenges the readers may have and provide counterarguments or mitigation strategies.
- Recap the main points: Summarize the key points you’ve discussed in the proposal.
- Reinforce the benefits: Emphasize the positive outcomes, benefits, or impact your proposal will have.
- Call to action: Clearly state what action you want the readers to take, such as approving the proposal, providing funding, or collaborating with you.

Review and Revise
- Proofread for clarity and coherence: Check for grammar, spelling, and punctuation errors.
- Ensure a logical flow: Read through your proposal to ensure the ideas are presented in a logical order and are easy to follow.
- Revise and refine: Fine-tune your proposal to make it concise, persuasive, and compelling.
Add Supplementary Materials
- Attach relevant documents: Include any supporting materials that strengthen your proposal, such as research findings, charts, graphs, or testimonials.
- Appendices: Add any additional information that might be useful but not essential to the main body of the proposal.
Formatting and Presentation
- Follow the guidelines: Adhere to any specific formatting guidelines provided by the organization or institution to which you are submitting the proposal.
- Use a professional tone and language: Ensure that your proposal is written in a clear, concise, and professional manner.
- Use headings and subheadings: Organize your proposal with clear headings and subheadings to improve readability.
- Pay attention to design: Use appropriate fonts, font sizes, and formatting styles to make your proposal visually appealing.
- Include a cover page: Create a cover page that includes the title of your proposal, your name or organization, the date, and any other required information.
Seek Feedback
- Share your proposal with trusted colleagues or mentors and ask for their feedback. Consider their suggestions for improvement and incorporate them into your proposal if necessary.
Finalize and Submit
- Make any final revisions based on the feedback received.
- Ensure that all required sections, attachments, and documentation are included.
- Double-check for any formatting, grammar, or spelling errors.
- Submit your proposal within the designated deadline and according to the submission guidelines provided.
Proposal Format
The format of a proposal can vary depending on the specific requirements of the organization or institution you are submitting it to. However, here is a general proposal format that you can follow:
1. Title Page:
- Include the title of your proposal, your name or organization’s name, the date, and any other relevant information specified by the guidelines.
2. Executive Summary:
- Provide a concise overview of your proposal, highlighting the key points and objectives.
- Summarize the problem, proposed solution, and anticipated benefits.
- Keep it brief and engaging, as this section is often read first and should capture the reader’s attention.
3. Introduction:
- State the problem or issue you are addressing and its significance.
- Provide background information to help the reader understand the context and importance of the problem.
- Clearly state the purpose and objectives of your proposal.
4. Problem Statement:
- Describe the problem in detail, highlighting its impact and consequences.
- Use data, statistics, or examples to support your claims and demonstrate the need for a solution.
5. Proposed Solution or Project Description:
- Explain your proposed solution or project in a clear and detailed manner.
- Describe how your solution addresses the problem and why it is the most effective approach.
- Include information on the methods, strategies, or activities you will undertake to implement your solution.
- Highlight any unique features, innovations, or advantages of your proposal.
6. Methodology:
- Provide a step-by-step explanation of the methodology or approach you will use to implement your proposal.
- Include a timeline or schedule that outlines the key milestones, tasks, and deliverables.
- Clearly describe the resources, personnel, or expertise required for each phase of the project.
7. Evaluation and Success Metrics:
- Explain how you will measure the success or effectiveness of your proposal.
- Identify specific metrics, indicators, or evaluation methods that will be used.
- Describe how you will track progress, gather feedback, and make adjustments as needed.
- Present a detailed budget that outlines the financial resources required for your proposal.
- Include all relevant costs, such as personnel, materials, equipment, and any other expenses.
- Provide a justification for each item in the budget.
9. Conclusion:
- Summarize the main points of your proposal.
- Reiterate the benefits and positive outcomes of implementing your proposal.
- Emphasize the value and impact it will have on the organization or community.
10. Appendices:
- Include any additional supporting materials, such as research findings, charts, graphs, or testimonials.
- Attach any relevant documents that provide further information but are not essential to the main body of the proposal.
Proposal Template
Here’s a basic proposal template that you can use as a starting point for creating your own proposal:
Dear [Recipient’s Name],
I am writing to submit a proposal for [briefly state the purpose of the proposal and its significance]. This proposal outlines a comprehensive solution to address [describe the problem or issue] and presents an actionable plan to achieve the desired objectives.
Thank you for considering this proposal. I believe that implementing this solution will significantly contribute to [organization’s or community’s goals]. I am available to discuss the proposal in more detail at your convenience. Please feel free to contact me at [your email address or phone number].
Yours sincerely,
Note: This template is a starting point and should be customized to meet the specific requirements and guidelines provided by the organization or institution to which you are submitting the proposal.
Proposal Sample
Here’s a sample proposal to give you an idea of how it could be structured and written:
Subject : Proposal for Implementation of Environmental Education Program
I am pleased to submit this proposal for your consideration, outlining a comprehensive plan for the implementation of an Environmental Education Program. This program aims to address the critical need for environmental awareness and education among the community, with the objective of fostering a sense of responsibility and sustainability.
Executive Summary: Our proposed Environmental Education Program is designed to provide engaging and interactive educational opportunities for individuals of all ages. By combining classroom learning, hands-on activities, and community engagement, we aim to create a long-lasting impact on environmental conservation practices and attitudes.
Introduction: The state of our environment is facing significant challenges, including climate change, habitat loss, and pollution. It is essential to equip individuals with the knowledge and skills to understand these issues and take action. This proposal seeks to bridge the gap in environmental education and inspire a sense of environmental stewardship among the community.
Problem Statement: The lack of environmental education programs has resulted in limited awareness and understanding of environmental issues. As a result, individuals are less likely to adopt sustainable practices or actively contribute to conservation efforts. Our program aims to address this gap and empower individuals to become environmentally conscious and responsible citizens.
Proposed Solution or Project Description: Our Environmental Education Program will comprise a range of activities, including workshops, field trips, and community initiatives. We will collaborate with local schools, community centers, and environmental organizations to ensure broad participation and maximum impact. By incorporating interactive learning experiences, such as nature walks, recycling drives, and eco-craft sessions, we aim to make environmental education engaging and enjoyable.
Methodology: Our program will be structured into modules that cover key environmental themes, such as biodiversity, climate change, waste management, and sustainable living. Each module will include a mix of classroom sessions, hands-on activities, and practical field experiences. We will also leverage technology, such as educational apps and online resources, to enhance learning outcomes.
Evaluation and Success Metrics: We will employ a combination of quantitative and qualitative measures to evaluate the effectiveness of the program. Pre- and post-assessments will gauge knowledge gain, while surveys and feedback forms will assess participant satisfaction and behavior change. We will also track the number of community engagement activities and the adoption of sustainable practices as indicators of success.
Budget: Please find attached a detailed budget breakdown for the implementation of the Environmental Education Program. The budget covers personnel costs, materials and supplies, transportation, and outreach expenses. We have ensured cost-effectiveness while maintaining the quality and impact of the program.
Conclusion: By implementing this Environmental Education Program, we have the opportunity to make a significant difference in our community’s environmental consciousness and practices. We are confident that this program will foster a generation of individuals who are passionate about protecting our environment and taking sustainable actions. We look forward to discussing the proposal further and working together to make a positive impact.
Thank you for your time and consideration. Should you have any questions or require additional information, please do not hesitate to contact me at [your email address or phone number].
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Grant Proposal – Example, Template and Guide

How To Write A Business Proposal – Step-by-Step...

Business Proposal – Templates, Examples and Guide

How To Write A Research Proposal – Step-by-Step...

Proposal – Types, Examples, and Writing Guide

How to choose an Appropriate Method for Research?
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Indian J Anaesth
- v.60(9); 2016 Sep
How to write a research proposal?
Department of Anaesthesiology, Bangalore Medical College and Research Institute, Bengaluru, Karnataka, India
Devika Rani Duggappa
Writing the proposal of a research work in the present era is a challenging task due to the constantly evolving trends in the qualitative research design and the need to incorporate medical advances into the methodology. The proposal is a detailed plan or ‘blueprint’ for the intended study, and once it is completed, the research project should flow smoothly. Even today, many of the proposals at post-graduate evaluation committees and application proposals for funding are substandard. A search was conducted with keywords such as research proposal, writing proposal and qualitative using search engines, namely, PubMed and Google Scholar, and an attempt has been made to provide broad guidelines for writing a scientifically appropriate research proposal.
INTRODUCTION
A clean, well-thought-out proposal forms the backbone for the research itself and hence becomes the most important step in the process of conduct of research.[ 1 ] The objective of preparing a research proposal would be to obtain approvals from various committees including ethics committee [details under ‘Research methodology II’ section [ Table 1 ] in this issue of IJA) and to request for grants. However, there are very few universally accepted guidelines for preparation of a good quality research proposal. A search was performed with keywords such as research proposal, funding, qualitative and writing proposals using search engines, namely, PubMed, Google Scholar and Scopus.
Five ‘C’s while writing a literature review

BASIC REQUIREMENTS OF A RESEARCH PROPOSAL
A proposal needs to show how your work fits into what is already known about the topic and what new paradigm will it add to the literature, while specifying the question that the research will answer, establishing its significance, and the implications of the answer.[ 2 ] The proposal must be capable of convincing the evaluation committee about the credibility, achievability, practicality and reproducibility (repeatability) of the research design.[ 3 ] Four categories of audience with different expectations may be present in the evaluation committees, namely academic colleagues, policy-makers, practitioners and lay audiences who evaluate the research proposal. Tips for preparation of a good research proposal include; ‘be practical, be persuasive, make broader links, aim for crystal clarity and plan before you write’. A researcher must be balanced, with a realistic understanding of what can be achieved. Being persuasive implies that researcher must be able to convince other researchers, research funding agencies, educational institutions and supervisors that the research is worth getting approval. The aim of the researcher should be clearly stated in simple language that describes the research in a way that non-specialists can comprehend, without use of jargons. The proposal must not only demonstrate that it is based on an intelligent understanding of the existing literature but also show that the writer has thought about the time needed to conduct each stage of the research.[ 4 , 5 ]
CONTENTS OF A RESEARCH PROPOSAL
The contents or formats of a research proposal vary depending on the requirements of evaluation committee and are generally provided by the evaluation committee or the institution.
In general, a cover page should contain the (i) title of the proposal, (ii) name and affiliation of the researcher (principal investigator) and co-investigators, (iii) institutional affiliation (degree of the investigator and the name of institution where the study will be performed), details of contact such as phone numbers, E-mail id's and lines for signatures of investigators.
The main contents of the proposal may be presented under the following headings: (i) introduction, (ii) review of literature, (iii) aims and objectives, (iv) research design and methods, (v) ethical considerations, (vi) budget, (vii) appendices and (viii) citations.[ 4 ]
Introduction
It is also sometimes termed as ‘need for study’ or ‘abstract’. Introduction is an initial pitch of an idea; it sets the scene and puts the research in context.[ 6 ] The introduction should be designed to create interest in the reader about the topic and proposal. It should convey to the reader, what you want to do, what necessitates the study and your passion for the topic.[ 7 ] Some questions that can be used to assess the significance of the study are: (i) Who has an interest in the domain of inquiry? (ii) What do we already know about the topic? (iii) What has not been answered adequately in previous research and practice? (iv) How will this research add to knowledge, practice and policy in this area? Some of the evaluation committees, expect the last two questions, elaborated under a separate heading of ‘background and significance’.[ 8 ] Introduction should also contain the hypothesis behind the research design. If hypothesis cannot be constructed, the line of inquiry to be used in the research must be indicated.
Review of literature
It refers to all sources of scientific evidence pertaining to the topic in interest. In the present era of digitalisation and easy accessibility, there is an enormous amount of relevant data available, making it a challenge for the researcher to include all of it in his/her review.[ 9 ] It is crucial to structure this section intelligently so that the reader can grasp the argument related to your study in relation to that of other researchers, while still demonstrating to your readers that your work is original and innovative. It is preferable to summarise each article in a paragraph, highlighting the details pertinent to the topic of interest. The progression of review can move from the more general to the more focused studies, or a historical progression can be used to develop the story, without making it exhaustive.[ 1 ] Literature should include supporting data, disagreements and controversies. Five ‘C's may be kept in mind while writing a literature review[ 10 ] [ Table 1 ].
Aims and objectives
The research purpose (or goal or aim) gives a broad indication of what the researcher wishes to achieve in the research. The hypothesis to be tested can be the aim of the study. The objectives related to parameters or tools used to achieve the aim are generally categorised as primary and secondary objectives.
Research design and method
The objective here is to convince the reader that the overall research design and methods of analysis will correctly address the research problem and to impress upon the reader that the methodology/sources chosen are appropriate for the specific topic. It should be unmistakably tied to the specific aims of your study.
In this section, the methods and sources used to conduct the research must be discussed, including specific references to sites, databases, key texts or authors that will be indispensable to the project. There should be specific mention about the methodological approaches to be undertaken to gather information, about the techniques to be used to analyse it and about the tests of external validity to which researcher is committed.[ 10 , 11 ]
The components of this section include the following:[ 4 ]
Population and sample
Population refers to all the elements (individuals, objects or substances) that meet certain criteria for inclusion in a given universe,[ 12 ] and sample refers to subset of population which meets the inclusion criteria for enrolment into the study. The inclusion and exclusion criteria should be clearly defined. The details pertaining to sample size are discussed in the article “Sample size calculation: Basic priniciples” published in this issue of IJA.
Data collection
The researcher is expected to give a detailed account of the methodology adopted for collection of data, which include the time frame required for the research. The methodology should be tested for its validity and ensure that, in pursuit of achieving the results, the participant's life is not jeopardised. The author should anticipate and acknowledge any potential barrier and pitfall in carrying out the research design and explain plans to address them, thereby avoiding lacunae due to incomplete data collection. If the researcher is planning to acquire data through interviews or questionnaires, copy of the questions used for the same should be attached as an annexure with the proposal.
Rigor (soundness of the research)
This addresses the strength of the research with respect to its neutrality, consistency and applicability. Rigor must be reflected throughout the proposal.
It refers to the robustness of a research method against bias. The author should convey the measures taken to avoid bias, viz. blinding and randomisation, in an elaborate way, thus ensuring that the result obtained from the adopted method is purely as chance and not influenced by other confounding variables.
Consistency
Consistency considers whether the findings will be consistent if the inquiry was replicated with the same participants and in a similar context. This can be achieved by adopting standard and universally accepted methods and scales.
Applicability
Applicability refers to the degree to which the findings can be applied to different contexts and groups.[ 13 ]
Data analysis
This section deals with the reduction and reconstruction of data and its analysis including sample size calculation. The researcher is expected to explain the steps adopted for coding and sorting the data obtained. Various tests to be used to analyse the data for its robustness, significance should be clearly stated. Author should also mention the names of statistician and suitable software which will be used in due course of data analysis and their contribution to data analysis and sample calculation.[ 9 ]
Ethical considerations
Medical research introduces special moral and ethical problems that are not usually encountered by other researchers during data collection, and hence, the researcher should take special care in ensuring that ethical standards are met. Ethical considerations refer to the protection of the participants' rights (right to self-determination, right to privacy, right to autonomy and confidentiality, right to fair treatment and right to protection from discomfort and harm), obtaining informed consent and the institutional review process (ethical approval). The researcher needs to provide adequate information on each of these aspects.
Informed consent needs to be obtained from the participants (details discussed in further chapters), as well as the research site and the relevant authorities.
When the researcher prepares a research budget, he/she should predict and cost all aspects of the research and then add an additional allowance for unpredictable disasters, delays and rising costs. All items in the budget should be justified.
Appendices are documents that support the proposal and application. The appendices will be specific for each proposal but documents that are usually required include informed consent form, supporting documents, questionnaires, measurement tools and patient information of the study in layman's language.
As with any scholarly research paper, you must cite the sources you used in composing your proposal. Although the words ‘references and bibliography’ are different, they are used interchangeably. It refers to all references cited in the research proposal.
Successful, qualitative research proposals should communicate the researcher's knowledge of the field and method and convey the emergent nature of the qualitative design. The proposal should follow a discernible logic from the introduction to presentation of the appendices.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
A conceptual framework proposed through literature review to determine the dimensions of social transparency in global supply chains
- Published: 16 May 2024
Cite this article

- Preethi Raja 1 &
- Usha Mohan ORCID: orcid.org/0000-0003-2161-7600 1
The current focus in supply chain management (SCM) research revolves around the relationship between sustainability and supply chain transparency (SCT). Despite the three pillars of sustainability – environmental, social, and economic- the limited and scattered analysis is on the social part, and the least is on socially responsible supply chain management (SR-SCM). SCT plays a significant role in elevating the sustainability of the supply chain. This review paper emphasizes the integration of SCT and sustainable supply chain, especially the social aspect as SR-SCM, and coining the new term social transparency (ST). ST is openness to communicating details about the impact of business on people, their well-being, and compliance with social sustainability standards and policies. This paper establishes a conceptual framework using three research methods. systematic literature review, content analysis-based literature review, and framework development. By locating studies in databases like EBSCO, Scopus, and Web of Science, 273 peer-reviewed articles were identified in the intersection of social sustainability, supply chains, and transparency. Finally, the framework proposes five dimensions: tracking and tracing suppliers till provenance, product and process specifications, financial transaction information, social sustainability policies and compliance, and performance assessment to determine ST in global supply chains.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
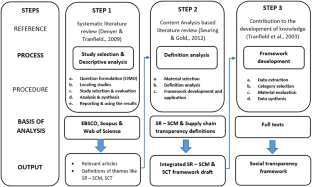
Data availability
The data that supports the findings of this systematic literature review and content analysis are either included in this manuscript or are publicly available in the referenced sources. All included studies and their respective citations are provided in the reference section. Any additional data or materials used for this review can be obtained upon request from the corresponding author.
Abbreviations
Supply chain management
Socially responsible supply chain management
Supply Chain Transparency
Social Transparency
Multinational Corporations
Code of Conduct
Corporate Social Responsibility
Preferred Reporting Items for Systematic Reviews and Meta-analyses
Radio frequency Identification
Internet of Things
Sustainable Supply Chain Management
Supply Chain
Textile Standard Certification
Worldwide Responsible Accredited Production
Global Organic Textile Standard
Global Recycled Standard
Registration, Evaluation, Authorization and Restriction on the use of Chemicals
Social Accountability International Certification
Indian Standards Institution Mark
Bureau of Indian Standards
Abdul S, Khan R, Zkik K, Belhadi A, Kamble SS (2021) Evaluating barriers and solutions for social sustainability adoption in multi-tier supply chains. Int J Prod Res 0(0):1–20. https://doi.org/10.1080/00207543.2021.1876271
Article Google Scholar
Al-Khatib AW (2023) Internet of things, big data analytics and operational performance: the mediating effect of supply chain visibility. J Manuf Technol Manage 34(1):1–24. https://doi.org/10.1108/JMTM-08-2022-0310
Awaysheh A, Klassen RD (2010) The impact of supply chain structure on the use of supplier socially responsible practices. Int J Oper Prod Manage, 30 (12)
Bangladesh garment workers (2023) ‘frustrated’ by Gov’t wage hike after protests. The minimum wage increase comes after weeks of the worst protests in a decade hit Bangladesh’s major industrial areas. November 08, [Cited 30.01.2024]. https://www.aljazeera.com/news/2023/11/8/bangladesh-garment-workers-frustrated-by-govts-wage-hike-after-protests#:~:text=Bangladesh&20is&20the&20second&20largest,according&20to&20the&20manufacturers'&20association .
Bangladesh workers’ protest (2023) 150 factories shut, cases against 11k workers. Bangladesh’s garment workers earn $95 a month as minimum wage and are demanding a minimum wage of $208 a month. BS Web Team November 13, [Cited 30.01.2024]. https://www.business-standard.com/world-news/bangladesh-workers-protest-150-factories-shut-cases-against-11k-workers-123111300291_1.html
Bozic D (2015) From Haute Couture to Fast-Fashion: Evaluating Social Transparency in Global Apparel Supply Chains. MIT Thesis
Brun A, Karaosman H, Barresi T (2020) Supply chain collaboration for transparency. Sustain (Switzerland), 12 (11)
Carter CR, Rogers DS (2008) A framework of sustainable supply chain management: moving toward new theory. Int J Phys Distribution Logistics Manage
Delaney A, Connor T (2016) Forced Labour in the Textile and Garment Sector in Tamil Nadu, South India Strategies for Redress . October , 1–67. http://corporateaccountabilityresearch.net/njm-report-xiii-sumangali
Denyer D, Tranfield D (2009) Producing a systematic review. In: Buchanan DA, Bryman A (eds) The sage handbook of organizational research methods. Sage Publications Ltd., pp 671–689
Doorey DJ, Doorey DJ (2018) The transparent supply chain: from resistance to implementation at Nike. J Bus Ethics 103(4):587–603
Egels-Zandén N, Hansson N (2016) Supply Chain transparency as a consumer or corporate Tool: the case of Nudie Jeans Co. J Consum Policy 39(4):377–395. https://doi.org/10.1007/s10603-015-9283-7
Egels-Zandén N, Hulthén K, Wulff G (2015) Trade-offs in supply chain transparency: the case of Nudie Jeans Co. J Clean Prod 107:95–104
Elkington J (1998) Accounting for the triple bottom line. Measuring Business Excellence
Fair Labor Association (2012) Understanding the Characteristics of the Sumangali Scheme in Tamil Nadu Textile & Garment Industry and Supply Chain Linkages . May . www.fairlabor.orgwww.solidaridadnetwork.org
Faisal M, Sabir N, Bin L (2023) Operationalizing transparency operationalizing in supply chains using a systematic literature review and graph theoretic approach. https://doi.org/10.1108/BIJ-05-2022-0291
Francisco K, Swanson D (2018) The Supply Chain Has No Clothes: Technology Adoption of Blockchain for Supply Chain Transparency. Logistics
Fraser IJ, Müller M, Schwarzkopf J (2020) Transparency for multi-tier sustainable supply chain management: a case study of a multi-tier transparency approach for SSCM in the automotive industry. Sustain (Switzerland) 12(5):1–24
Google Scholar
Hohn MM, Durach CF (2023) Taking a different view: theorizing on firms ’ development toward an integrative view on socially sustainable supply chain management . 53 (1), 13–34. https://doi.org/10.1007/s10551-012-1245-2
https://cleanclothes.org/news/2015/03/18/rana-plaza-survivor-and-others-arrested-at-childrens-place-headquarters
https://cleanclothes.org/campaigns/the-accord
Huq FA, Stevenson M, Zorzini M (2014) Social sustainability in developing country suppliers an exploratory study in the ready made garments industry of Bangladesh. Int J Oper Prod Manage
Jamalnia A, Gong Y, Govindan K (2023) Sub-supplier’s sustainability management in multi-tier supply chains: a systematic literature review on the contingency variables, and a conceptual framework. Int J Prod Econ 255(January 2022):108671. https://doi.org/10.1016/j.ijpe.2022.108671
Klassen RD, Vereecke A (2012) Social issues in supply chains: capabilities link responsibility, risk (opportunity), and performance. Intern J Prod Econ
Kraft T, Valdés L, Zheng Y (2018) Supply Chain visibility and social responsibility: investigating consumers ’ behaviors and motives. Manufacturing & Service Operations Management
Lamming RC, Caldwell ND, Harrison DA, Phillips W (2001) Transparency in Supply Relationships: Concept and Practice . November , 4–10
Limited KEDSP (2019) Evaluation of Sumangali_Eradication of extremely exploitative working conditions in Southern India’s textile industry . June
Mayer DM, Ong M, Sonenshein S, Ashford S (2019) To Get Companies to Take Action on Social Issues, Emphasize Morals, Not the Business Case. [Website]. [Cited 11.11.2023]. Available: https://hbr.org/2019/02/to-get-companies-to-take-action-on-social-issues-emphasize-morals-not-the-business-case
McGrath P, McCarthy L, Marshall D, Rehme J (2021) Tools and technologies of transparency in sustainable global supply chains. Calif Manag Rev 64(1):67–89
Moher D, Liberati A, Tetzlaff J, Altman DG, Antes G, Atkins D, Barbour V, Barrowman N, Berlin JA, Clark J, Clarke M, Cook D, D’Amico R, Deeks JJ, Devereaux PJ, Dickersin K, Egger M, Ernst E, Gøtzsche PC, Tugwell P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6(7). https://doi.org/10.1371/journal.pmed.1000097
Montecchi M, Plangger K, C. West D (2021) Supply chain transparency: a bibliometric review and research agenda. Int J Prod Econ 238(August 2020):108152. https://doi.org/10.1016/j.ijpe.2021.108152
Morgan TR, Gabler CB, Manhart PS (2023) Supply chain transparency: theoretical perspectives for future research. The International Journal of Logistics Management , 2008 . https://doi.org/10.1108/ijlm-02-2021-0115
Parmar BL, Freeman RE, Harrison JS, Wicks AC, Purnell L, Bidhan L, Edward R, Jeffrey S, Andrew C (2011) The Academy of Management annals Stakeholder Theory: the state of the art Stakeholder Theory : the state of the art. Management 936836193:403–445
Report of the World Commission on Environment and Development (1987) United Nations. Our Common Future
Robledo P, Triebich M (2020) Position Paper on Transparency. Clean Clothes Campaign , April . www.fashionchecker.org
Rodr JA, Enez CGIM, Ramon E, Pagell M (2016) NGO’s initiatives to enhance social sustainability in the supply chain: poverty alleviation through supplier development programs. J Supply Chain Manage, 1–26
Sancha C, Gimenez C, Sierra V (2016) Achieving a socially responsible supply chain through assessment and collaboration. J Clean Prod, 112
Schäfer N (2022) Making transparency transparent: a systematic literature review to define and frame supply chain transparency in the context of sustainability. Manage Rev Q
Senyo PK, Osabutey ELC (2023) Transdisciplinary perspective on sustainable multi-tier supply chains: a triple bottom line inspired framework and future research directions. Int J Prod Res 61(14):4918–4933. https://doi.org/10.1080/00207543.2021.1946194
Seuring S, Gold S (2012) Conducting content-analysis based literature reviews in supply chain management. Supply Chain Manage 17(5):544–555
Shrivastava P, Hart SL (1995) Creating sustainable corporations. Bus Strategy Environ 4:154–165
Sikdar SK (2003) Sustainability development and sustainability metrics. Am Inst Chem Eng, 49 (8)
Sodhi MS, Tang CS (2019) Research Opportunities in Supply Chain Transparency . 0 (0), 1–14. https://doi.org/10.1111/poms.13115
Spence L, Bourlakis M (2009) The evolution from corporate social responsibility to supply chain responsibility: the case of Waitrose. Supply Chain Management: Int J
Tranfield D, Denyer D, Smart P (2003) Towards a methodology for developing evidence-informed management knowledge by means of systematic review. Br J Manag 14:207–222
Venkatesh VG, Kang K, Wang B, Zhong RY, Zhang A (2020a) System architecture for blockchain based transparency of supply chain social sustainability. Robotics and Computer-Integrated Manufacturing
Venkatesh VG, Zhang A, Deakins E, Venkatesh M (2020b) Drivers of sub-supplier social sustainability compliance: an emerging economy perspective. Supply Chain Management: Int J
Wognum PMN, Bremmers H, Trienekens JH, Vorst JGA, Van Der J, Bloemhof JM (2011) Systems for sustainability and transparency of food supply chains – Current status and challenges. Advanced Engineering Informatics
Yawar SA, Seuring S (2015) Management of Social issues in Supply chains: a Literature Review Exploring Social Issues, actions and performance outcomes. J Bus Ethics
Download references
There is no funding for this project.
Author information
Authors and affiliations.
Department of Management Studies, IIT Madras, Chennai, 600036, Tamil Nadu, India
Preethi Raja & Usha Mohan
You can also search for this author in PubMed Google Scholar
Ethics declarations
Conflict of interest.
There is no conflict of interest in this paper.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, supplementary material 3, rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Raja, P., Mohan, U. A conceptual framework proposed through literature review to determine the dimensions of social transparency in global supply chains. Manag Rev Q (2024). https://doi.org/10.1007/s11301-024-00440-1
Download citation
Received : 21 July 2023
Accepted : 26 April 2024
Published : 16 May 2024
DOI : https://doi.org/10.1007/s11301-024-00440-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Social sustainability
- Socially responsible supply chain
- Supply chain transparency
- Social transparency evaluation framework
- Corporate social responsibility
- Conceptual framework development
- Find a journal
- Publish with us
- Track your research
- Open access
- Published: 13 May 2024
Neighborhood based computational approaches for the prediction of lncRNA-disease associations
- Mariella Bonomo 1 &
- Simona E. Rombo 1 , 2
BMC Bioinformatics volume 25 , Article number: 187 ( 2024 ) Cite this article
106 Accesses
Metrics details
Long non-coding RNAs (lncRNAs) are a class of molecules involved in important biological processes. Extensive efforts have been provided to get deeper understanding of disease mechanisms at the lncRNA level, guiding towards the detection of biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, due to costs and time complexity, the number of possible disease-related lncRNAs verified by traditional biological experiments is very limited. Computational approaches for the prediction of disease-lncRNA associations allow to identify the most promising candidates to be verified in laboratory, reducing costs and time consuming.
We propose novel approaches for the prediction of lncRNA-disease associations, all sharing the idea of exploring associations among lncRNAs, other intermediate molecules (e.g., miRNAs) and diseases, suitably represented by tripartite graphs. Indeed, while only a few lncRNA-disease associations are still known, plenty of interactions between lncRNAs and other molecules, as well as associations of the latters with diseases, are available. A first approach presented here, NGH, relies on neighborhood analysis performed on a tripartite graph, built upon lncRNAs, miRNAs and diseases. A second approach (CF) relies on collaborative filtering; a third approach (NGH-CF) is obtained boosting NGH by collaborative filtering. The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
Availability
Source code and sample datasets are available at: https://github.com/marybonomo/LDAsPredictionApproaches.git
Peer Review reports
Introduction
More than \(98\%\) of the human genome consists of non-coding regions, considered in the past as “junk” DNA. However, in the last decades evidence has been shown that non-coding genome elements often play an important role in regulating various critical biological processes [ 1 ]. An important class of non-coding molecules which have started to receive great attention in the last few years is represented by long non-coding RNAs (lncRNAs), that is, RNAs not translated into functional proteins, and longer than 200 nucleotides.
LncRNAs have been found to interplay with other molecules in order to perform important biological tasks, such as modulating chromatin function, regulating the assembly and function of membraneless nuclear bodies, interfering with signalling pathways [ 2 , 3 ]. Many of these functions ultimately affect gene expression in diverse biological and physiopathological contexts, such as in neuronal disorders, immune responses and cancer. Therefore, the alteration and dysregulation of lncRNAs have been associated with the occurrence and progress of many complex diseases [ 4 ].
The discovery of novel lncRNA-disease associations (LDAs) may provide valuable input to the understanding of disease mechanisms at lncRNA level, as well as to the detection of disease biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, verifying that a specific lncRNA may have a role in the occurrence/progress of a given disease is an expensive process, therefore the number of disease-related lncRNAs verified by traditional biological experiments is yet very limited. Computational approaches for the prediction of potential LDAs can effectively decrease the time and cost of biological experiments, allowing for the identification of the most promising lncRNA-disease pairs to be further verified in laboratory (see [ 5 ] for a comprehensive review on the topic). Such approaches often train predictive models on the basis of the known and experimentally validated lncRNA-disease pairs (e.g., [ 6 , 7 , 8 , 9 ]). In other cases, they rely on the analysis of lncRNAs related information stored in public databases, such as their interaction with other types of molecules (e.g., [ 10 , 11 , 12 , 13 , 14 , 15 ]). As an example, large amounts of lncRNA-miRNA interactions have been collected in public databases, and plenty of experimentally confirmed miRNA-disease associations are available as well. However, although non-coding RNA function and its association with human complex diseases have been widely studied in the literature (see [ 16 , 17 , 18 ]), how to provide biologists with more accurate and ready-to-use software tools for LDAs prediction is yet an open challenge, due to the specific characteristics of lncRNAs (e.g., they are much less characterized than other non-coding RNAs.)
We propose three novel computational approaches for the prediction of LDAs, relying on the use of known lncRNA-miRNA interactions (LMIs) and miRNA-disease associations (MDAs). In particular, we model the problem of LDAs prediction as a neighborhood analysis performed on tripartite graphs, where the three sets of vertices represent lncRNAs, miRNAs and diseases, respectively, and vertices are linked according to LMIs and MDAs. Based on the assumption that similar lncRNAs interact with similar diseases [ 12 ], the first approach proposed here (NGH) aims at identifying novel LDAs by analyzing the behaviour of lncRNAs which are neighbors , in terms of their intermediate relationships with miRNAs. The main idea here is that neighborhood analysis automatically guides towards the detection of similar behaviours, and without the need of using a-priory known LDAs for training. Therefore, differently than other approaches from the literature, those proposed here do not involve verified LDAs in the prediction step, thus avoiding possible biases due to the fact that the number and variety of verified LDAs is yet very limited. The second presented approach (CF) relies on collaborative filtering, applied on the basis of common miRNAs shared by different lncRNAs. We have also explored the combination of neighborhood analysis with collaborative filtering, showing that this notably improves the LDAs prediction accuracy. Indeed, the third approach we have designed (NGH-CF) boosts NGH with collaborative filtering, and it is the best performing one, although also NGH and CF have been able to reach high accuracy values across all the different considered validation tests. In particular, Fig. 1 summarizes the research flowchart explained above.
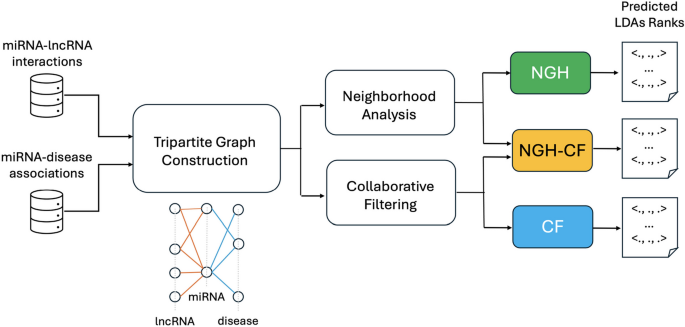
Flowchart of the research pipeline. The miRNA-lncRNA interactions and miRNA-disease associations are exploited for the construction of the tripartite graph. The tripartite graph, in its turn, is at the basis of both neighborhood analysis and collaborative filtering steps, from which the three proposed approaches are obtained: NGH from neighborhood analysis, CF from collaborative filtering, NGH-CF from the combination of the two ones. Each prediction approach returns in output a LDAs rank
The proposed approaches have been exhaustively validated on both synthetic and real datasets, and the result is that they outperform (also significantly) the other methods from the literature. The experimental analysis shows that the improvement in accuracy achieved by the methods proposed here is due to their ability in capturing specific situations neglected by competitors. Examples of that are represented by true LDAs, detected by our approaches and not by the other approaches in the literature, where the involved lncRNA does not present intermediate molecules in common with the associated disease, although its neighbor lncRNAs share a large number of miRNAs with that disease. Moreover, it is shown that our approaches are robust to noise obtained by perturbing a controlled percentage of lncRNA-miRNA interactions and miRNA-disease associations, with NGH-CF the best one also for robustness. The obtained experimental results show that the prediction methods proposed here may effectively support biologists in selecting significant associations to be further verified in laboratory.
Novel putative LDAs coming from the consensus of the three proposed methods, and not yet registered in the available databases as experimentally verified, are provided. Interestingly, the core of novel LDAs returned with highest score by all three approaches finds evidence in the recent literature, while many other high scored predicted LDAs involve less studied lncRNAs, thus providing useful insights for their better characterization.
A first group of approaches aim at using existing true validated cases to train the prediction system, in order to make it able to correctly detect novel cases.
In [ 19 ] a Laplacian Regularized Least Squares is proposed to infer candidates LDAs ( LRLSLDA ) by applying a semi-supervised learning framework. LRLSLDA assumes that similar diseases tend to correlate with functionally similar lncRNAs, and vice versa. Thus, known LDAs and lncRNA expression profiles are combined to prioritize disease-associated lncRNA candidates by LRLSLDA, which does not require negative samples (i.e., confirmed uncorrelated LDAs). In [ 20 ] the method SKF-LDA is proposed that constructs a lncRNA-disease correlation matrix, based on the known LDAs. Then, it calculates the similarity between lncRNAs and that between diseases, according to specific metrics, and integrates such data. Finally, a predicted LDA matrix is obtained by the Laplacian Regularized Least Squares method. The method ENCFLDA [ 6 ] combines matrix decomposition and collaborative filtering. It uses matrix factorization combined with elastic networks and a collaborative filtering algorithm, making the prediction model more stable and eliminating the problem of data over-fitting. HGNNLDA recently proposed in [ 21 ] is based on hypergraph neural network, where the associations are modeled as a lncRNA-drug bipartite graph to build lncRNA hypergraph and drug hypergraph. Hypergraph convolution is then used to learn correlation of higher-order neighbors from the lncRNA and drug hypergraphs. LDAI-ISPS proposed in [ 22 ] is a LDAs inference approach based on space projections of integrated networks, recostructing the disease (lncRNA) integrated similarities network via integrating multiple information, such as disease semantic similarities, lncRNA functional similarities, and known LDAs. A space projection score is finally obtained via vector projections of the weighted networks. In [ 7 ] a consensual prediction approach called HOPEXGB is presented, to identify disease-related miRNAs and lncRNAs by high-order proximity preserved embedding and extreme gradient boosting. The authors build a heterogeneous disease-miRNA-lncRNA (DML) information network by linking lncRNA, miRNA, and disease nodes based on their correlation, and generate a negative dataset based on the similarities between unknown and known associations, in order to reduce the false negative rate in the data set for model construction. The method MAGCNSE proposed in [ 23 ] builds multiple feature matrices based on semantic similarity and disease Gaussian interaction profile kernel similarity of both lncRNAs and diseases. MAGCNSE adaptively assigns weights to the different feature matrices built upon the lncRNAs and diseases similarities. Then, it uses a convolutional neural network to further extract features from multi-channel feature matrices, in order to obtain the final representations of lncRNAs and diseases that is used for the LDAs prediction task.
LDAFGAN [ 8 ] is a model designed for predicting associations between long non-coding RNAs (lncRNAs) and diseases. This method is based on a generative and a discriminative networks, typically implemented as multilayer fully connected neural networks, which generate synthetic data based on some underlying distribution. The generative and discriminative networks are trained together in an adversarial manner. The generative network tries to generate realistic representations of lncRNA-disease associations, while the discriminative network tries to distinguish between real and fake associations. This adversarial training process helps the generative network learn to generate more realistic associations. Once the model is trained, it can predict associations between new lncRNAs and diseases without requiring associated data for those specific lncRNAs. The model captures the data distribution during training, which enables it to make predictions even for unseen lncRNAs. The approach GCNFORMER [ 9 ] is based on graph convolutional network and transformer. First, it integrates the intraclass similarity and interclass connections between miRNAs, lncRNAs and diseases, building a graph adjacency matrix. Then, the method extracts the features between various nodes, by a graph convolutional network. To obtain the global dependencies between inputs and outputs, a transformer encoder with a multiheaded attention mechanism to forecast lncRNA-disease associations is finally applied.
As for the approaches summarized above, it is worth to point out that they may suffer of the fact that the experimentally verified LDAs are still very limited, therefore the training set may be rather incomplete and not enough diversified. For this reason, when such approaches are applied for de novo LDAs prediction, their performance may drastically go down [ 12 ].
Other approaches from the literature use intermediate molecules (e.g., miRNA) to infer novel LDAs. Such approaches are the most related to those we propose here.
The author in [ 11 ] proposes HGLDA , relying on HyperGeometric distribution for LDAs inference, that integrates MDAs and LMIs information. HGLDA has been successfully applied to predict Breast Cancer, Lung Cancer and Colorectal Cancer-related lncRNAs. NcPred [ 10 ] is a resource propagation technique, using a tripartite network where the edges associate each lncRNA with a disease through its targets. The algorithm proposed in [ 10 ] is based on a multilevel resource transfer technique, which computes the weights between each lncRNA-disease pair and, at each step, considers the resource transferred from the previous step. The approach in [ 24 ], referred to as LDA-TG for short in the following, is the antecedent of the approaches proposed here. It relies on the construction of a tripartite graph, built upon MDAs and LMIs. A score is assigned to each possible LDA ( l , d ) by considering both their respective interactions with common miRNAs, and the interactions with miRNAs shared by the considered disease d and other lncRNAs in the neighborhood of l on the tripartite graph. The approaches proposed here differ from LDA-TG for two main reasons. First, the score of LDA-TG is different from the one we introduce here, that allows to reach a better accuracy. Second, a further step based on collaborative filtering is considered here, which also improves the accuracy performance. A method for LDAs prediction relying on a matrix completion technique inspired by recommender systems is presented in [ 14 ]. A two-layer multi-weighted nearest-neighbor prediction model is adopted, using a method similar to memory-based collaborative filtering. Weights are assigned to neighbors for reassigning values to the target matrix, that is an adjacency matrix consisting of lncRNAs, diseases and miRNA. SSMF-BLNP [ 25 ] is based on the combination of selective similarity matrix fusion (SSMF) and bidirectional linear neighborhood label propagation (BLNP). In SSMF, self-similarity networks of lncRNAs and diseases are obtained by selective preprocessing and nonlinear iterative fusion. In BLNP, the initial LDAs are employed in both lncRNA and disease directions as label information for linear neighborhood label propagation.
A third category includes approaches based on integrative frameworks, proposed to take into account different types of information related to lncRNAs, such as their interactions with other molecules, their involvement in disorders and diseases, their similarities. This may improve the prediction step, taking into account simultaneously independent factors.
IntNetLncSim [ 26 ] relies on the construction of an integrated network that comprises lncRNA regulatory data, miRNA-mRNA and mRNA-mRNA interactions. The method computes a similarity score for all pairs of lncRNAs in the integrated network, then analyzes the information flow based on random walk with damping. This allows to infer novel LDAs by exploring the function of lncRNAs. SIMCLDA [ 12 ] identifies LDAs by using inductive matrix completion, based on the integration of known LDAs, disease-gene interactions and gene-gene interactions. The main idea in [ 12 ] is to extract feature vectors of lncRNAs and diseases by principal component analysis, and to calculate the interaction profile for a new lncRNA by the interaction profiles. MFLDA [ 27 ] is a Matrix Factorization based LDAs prediction model that first encodes directly (or indirectly) relevant data sources related to lncRNAs or diseases in individual relational data matrices, and presets weights for these matrices. Then, it simultaneously optimizes the weights and low-rank matrix tri-factorization of each relational data matrix. RWSF-BLP , proposed in [ 28 ], applies a random walk-based multi-similarity fusion method to integrate different similarity matrices, mainly based on semantic and expression data, and bidirectional label propagation. The framework LRWRHLDA is proposed in [ 15 ] based on the construction of a global multi-layer network for LDAs prediction. First, four isomorphic networks including a lncRNA similarity network, a disease similarity network, a gene similarity network and a miRNA similarity network are constructed. Then, six heterogeneous networks involving known lncRNA-disease, lncRNA-gene, lncRNA-miRNA, disease-gene, disease-miRNA, and gene-miRNA associations are built to design the multi-layer network. In [ 29 ] the LDAP-WMPS LDA prediction model is proposed, based on weight matrix and projection score. LDAP-WMPS consists on three steps: the first one computes the disease projection score; the second step calculates the lncRNA projection score; the third step fuses the disease projection score and the lncRNA projection score proportionally, then it normalizes them to get the prediction score matrix.
For most of the approaches summarized above, the performance is evaluated using the LOOCV framework, such that each known LDA is left out in turn as a test sample, and how well this test sample is ranked relative to the candidate samples (all the LDAs without the evidence to confirm their relationships) is computed.
The main goal of the research presented here is to provide more accurate computational methods for the prediction of novel LDAs, candidate for experimental validation in laboratory. To this aim, external information on both molecular interactions (e.g., lncRNA-miRNA interactions) and genotype-phenotype associations (e.g., miRNA-disease associations) is assumed to be available. Indeed, while only a restricted number of validated LDAs is yet available, large amounts of interactions between lncRNAs and other molecules (e.g., miRNAs, genes, proteins), as well as associations between these other molecules and diseases, are known and annotated in curated databases.
A commonly recognized assumption is that lncRNAs with similar behaviour in terms of their molecular interactions with other molecules, may also reflect such a similarity for their involvement in the occurrence and progress of disorders and diseases [ 12 ]. This is even more effective if the correlation with diseases is “mediated” by the molecules they interact with. Based on this observation, we have designed three novel prediction methods that all consider the notion of lncRNA “neighbors”, intended as lncRNAs which share common mediators among the molecules they physically interact with. Here, we focus on miRNAs as mediator molecules. However, the proposed approaches are general enough to allow also the inclusion of other different molecules. Relationships among lncRNAs, mediators and diseases are modeled through tripartite graphs in all the proposed approaches (see Fig. 1 that illustrates the flowchart of the presented research pipeline).
Problem statement Let \({\mathcal {L}}=\{l_1, l_2, \ldots , l_h\}\) be a set of lncRNAs and \({\mathcal {D}}=\{d_1, d_2, \ldots , d_k\}\) be a set of diseases. The goal is to return an ordered set of triplets \({\mathcal {R}}=\{\langle l_x, d_y, s_{xy}\rangle \}\) (with \(x\in [1,h]\) , and \(y\in [1,k]\) ), ranked according to the score \(s_{xy}\) .
The top triplets in \({\mathcal {R}}\) correspond to those pairs \((l_x, d_y)\) with most chances to represent putative LDAs which may be considered for further analysis in laboratory, while the triplets in the bottom correspond to lncRNAs and diseases which are unlikely to be related each other. A key aspect for the solution of the problem defined above is the score computation, that is the main aim of the approaches introduced in the following.
NGH: neighborhood based approach
A model of tripartite graph is adopted here to take into account that lncRNAs interacting with common mediators may be involved in common diseases.
Let \(T_{LMD}=\langle I, A \rangle\) be a tripartite graph defined on the three sets of disjoint vertexes L , M and D , such that \((l,m) \in I\) are edges between vertexes \(l \in L\) and \(m \in M\) , \((m,d) \in A\) are edges between vertexes \(m \in M\) and \(d \in D\) , respectively. In particular, L is associated to a set of lncRNAs, M to a set of miRNA and D to a set of diseases. Moreover, edges of the type ( l , m ) represent molecular interactions between lncRNAs and miRNA, experimentally validated in laboratory; edges of the type ( m , d ) correspond to known miRNA-disease associations, according to the existing literature. In both cases, interactions and associations annotated and stored in public databases may be taken into account.
The following definitions hold.
Definition 1
(Neighbors) Two lncRNAs \(l_h, l_k \in L\) are neighbors in \(T_{LMD}=\langle I, A \rangle\) if there exists at least a \(m_x \in M\) such that \((l_h, m_x) \in I\) and \((l_k, m_x) \in I\) .
Definition 2
(Prediction Score) The Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) and \(d_j \in D\) is defined as:
\(M_{l_i}\) is the set of annotated miRNA interacting with \(l_i\) ,
\(M_{d_j}\) is the set of miRNA found to be associated to \(d_j\) ,
\(M_{l_x}\) is the set of miRNA interacting with the neighbor \(l_x\) of \(l_i\) (for each neighbor of \(l_i\) ),
\(\alpha\) is a real value in [0, 1] used to balance the two terms of the formula.
Definition 3
(Normalized prediction score) The Normalized Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) , \(d_j \in D\) and \(s_{ij}\) is the Prediction Score for \((l_i,d_j)\) , is defined as:
NGH-CF: NGH extended with collaborative filtering
We remark that the main idea here is trying to infer the behaviour of a lncRNA, from that of its neighbors. Moreover, it is worth to point out that the notion of neighbor is related to the presence of miRNAs interacting with the same lncRNAs. However, not all the miRNA-lncRNA interactions have already been discovered, and miRNA-disease associations as well. This intuitively reminds to a typical context of data incompleteness where Collaborative Filtering may be successful in supporting the prediction process [ 30 ].
In more detail, what to be encoded by the Collaborative Filter is that lncRNAs presenting similar behaviours in terms of interactions with miRNAs, should reflect such a similarity also in their involvement with the occurrence and progress of diseases, mediated by those miRNAs. To this aim, a matrix R is considered here such that each element \(r_{ij}\) represents if (or to what extent) the lncRNA i and the disease j may be considered related. We call R relationship matrix (it is also known as rating matrix in other contexts, such as for example in the prediction of user-item associations). How to obtain \(r_{ij}\) is at the basis of the two variants of the approach presented in this section.
Due to the fact that R is usually a very sparse matrix, it can be factored into other two matrices L and D such that R \(\approx\) \(L\) \(^T\) \(D\) . In particular, matrix factorization models map both lncRNAs and diseases to a joint latent factor space F of dimensionality f , such that each lncRNA i is associated with a vector \(l_i \in F\) , each disease j with a vector \(d_j \in F\) , and their relationships are modeled as inner products in that space. Indeed, for each lncRNA i , the elements of \(l_i\) measure the extent to which it possesses those latent factors, and the same holds for each disease j and the corresponding elements of \(d_j\) . The resulting dot product in the factor space captures the affinity between lncRNA i and disease j , with reference to the considered latent factors. To this aim, there are two important tasks to be solved:
Mapping lncRNAs and diseases into the corresponding latent factors vectors.
Fill the matrix R , that is, the training set.
To learn the factor vectors \(l_i\) and \(d_j\) , a possible choice is to minimize the regularized squared error on the set of known relationships:
where \(\chi\) is the set of ( i , j ) pairs for which \(r_{ij}\) is not equal to zero in the matrix R . To this aim, we apply the ALS technique [ 31 ], which rotates between fixing the \(l_i\) ’s and fixing the \(d_j\) ’s. When all \(l_i\) ’s are fixed, the system recomputes the \(d_j\) ’s by solving a least-squares problem, and vice versa.
Filling the matrix R is performed according to two different criteria, resulting in the two different variants of the approach presented in this section, namely, CF and NGH-CF, respectively. According to the first criteria (CF), \(r_{ij}\) is set equal to 1 if the lncRNA i and the disease j share at least one miRNA in common, to 0 otherwise. The second variant (NGH-CF) works instead as a booster to improve the accuracy of NGH. In this latter case, the matrix R is filled by the normalized score ( 2 ). For both variants, the considered score to rank the predicted LDAs is given by the final value returned by the ALS technique applied on the corresponding matrix R .
Validation methodologies
We remark that the proposed approaches for LDAs prediction return a rank of LDAs, sorted according to the score that is characteristic of the considered approach, such that top triplets may be assumed as the most promising putative LDAs for further analysis in laboratory. As in other contexts [ 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 ], the performance of a prediction tool may be evaluated using suitable external criteria . Here, an external criterion relies on the existence of LDAs that are known to be true from the literature or, even better, from public repositories, where associations already verified in laboratory are annotated. A gold standard is constructed, containing only such true LDAs. The putative LDAs returned by the prediction method can thus be compared against those in the gold standard. In order to work properly, this validation methodology requires the gold standard information to be independent on that considered, in its turn, from the method under evaluation during its prediction task. This is satisfied in our case, due to the fact that all three approaches introduced in the previous sections do not exploit any type of knowledge referred to known LDAs during prediction, relying instead on known miRNA-lncRNA interactions and miRNA-disease associations, which come from independent sources.
According to the above mentioned validation methodology, the proposed approaches can be validated with references to the Receiver Operating Characteristics (ROC) analysis [ 34 ]. In particular, each predicted LDA is associated to a label, that is true if that association is contained in the considered gold standard, and false otherwise.
By varying the threshold value, it is possible to compute the true positive rate (TPR) and the false positive rate (FPR), by refferring to the percentage of the true/false predictions whose ranking is higher/below than the considered threshold value. ROC curve can be drawn by plotting TPR versus FPR at different threshold values. The Area Under ROC Curve (ROC-AUC) is further calculated to evaluate the performance of the tested methods. ROC-AUC equal to 1 indicates perfect performance, ROC-AUC equal to 0.5 random performance.
Similarly to the ROC curve, the Precision-Recall (PR) curve can be drawn as well, combining the positive predictive value (PPV, Precision), i.e., the fraction of predicted LDAs which are true in the gold standard, and the TPR (Recall), in a single visualization, at the threshold varying. The higher on y-axis the obtained curve is, the better the prediction method performance. The Area Under PR curve (AUPR) is more sensitive than AUC to the improvements for the positive class prediction [ 35 ], that is important for the case studied here. Indeed, only true LDAs are known, therefore no negative samples are included in the gold standard.
Another important measure useful to evaluate the prediction accuracy of a method and that can be considered here is the F1-score, defined as the harmonic mean of Precision and Recall to symmetrically represent both metrics in a single one.
We have validated the proposed approaches on both syntetic and real datasets, as explained below.
Synthetic data
A synthetic dataset has been built with 15 lncRNAs, 35 miRNA and 10 diseases, such that three different sets of LDAs may be identified, as follows (see also Table 1 , where the characteristics of each LDA are summarized).
Set 1: 26 LDAs, such that each lncRNA has from 3 to 4 miRNAs shared with the same disease (strongly linked lncRNAs) .
Set 2: 16 LDAs, each lncRNA having only one miRNA shared with a disease, and from 2 to 5 neighbors that are strongly linked with that same disease (directly linked lncRNAs and strong neighborhood) .
Set 3: 12 LDAs involving lncRNAs without any miRNA in common with a certain disease, and a number between 2 and 5 neighbors that are strongly linked with that same disease (only strong neighborhood) .
Experimentally verified data downloaded from starBase [ 36 ] and from HMDD [ 37 ] have been considered for the lncRNA-miRNA interactions and for the miRNA-disease associations, respectively. In particular, the latest version of HMDD, updated at 2019, has been used. Overall, \(1,\!114\) lncRNAs, \(1,\!058\) miRNAs, 885 diseases, \(10,\!112\) lncRNA-miRNA interactions and \(16,\!904\) miRNA-disease associations have been included in the analysis.
In order to evaluate the prediction accuracy of the approaches proposed here against those from the literature, three different gold standards have been considered. A first gold standard dataset GS1 has been obtained from the LncRNA-Disease database [ 38 ], resulting in 183 known and verified LDAs. A second, more restrictive, gold standard GS2 with 157 LDAs has been built by the intersection of data from [ 38 ] and [ 39 ]. Finally, also a larger gold standard dataset GS3 has been included in the analysis, by extracting LDAs from MNDRv2.0 database [ 40 ], where associations both experimentally verified and retrieved from manual literature curation are stored, resulting in 408 known LDAs.
Comparison on real data
The approaches proposed here have been compared against other approaches from the literature, over the three different gold standards described in the previous Section. In particular, all approaches considered from the literature have been run according to the default setting of their parameters, reported on the corresponding scientific publications and/or on their manual instructions.
Our approaches have been compared at first on GS1 against those approaches taking exactly the same input than ours, that are HGLDA [ 11 ], ncPred [ 10 ] and LDA-TG [ 24 ]. In particular, we have implemented HGLDA and used the corresponding p-value score, corrected by FDR as suggested by [ 11 ], for the ROC analysis. Moreover, we have normalized also the scores returned by ncPred and LDA-TG for the predicted LDAs, according to the formula in Definition 3 . Indeed, we have observed experimentally that such a normalization improves the accuracy of both methods from the literature, resulting in a better AUC. As for the novel approaches proposed here, the Normalized Prediction Score has been considered for NGH, while the approximated rating score resulting from ALS [ 31 ] is used for both CF and NGH-CF. Figure 2 shows the AUC scored by each method on GS1, while in Fig. 3 the different ROC curves are plotted. In particular, NGH scores a value of AUC equal to 0.914, thus outperforming the other three methods previously presented in the literature, i.e., HGLDA, ncPred and LDA-TG, that reach 0.876, 0.886 and 0.866, respectively (we remark also that performance of both ncPred and LDA-TG has been slightly improved with respect to their original one, by normalizing their scores). As for the novel approaches based on collaborative filtering, they both present a better accuracy than the others, with CF having AUC equal to 0.957 and NGH-CF to 0.966, respectively. Therefore, these results confirm that taking into account the collaborative effects of lncRNAs and miRNAs is useful to improve LDAs prediction, and the most successful approach is NGH-CF, that is, the neighborhood based approach boosted by collaborative filtering.
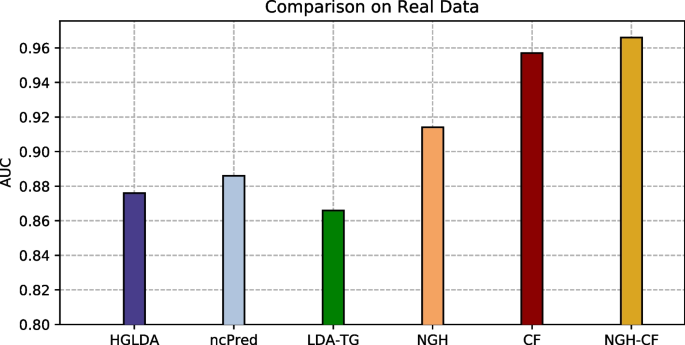
Comparison of the scored AUC on GS1
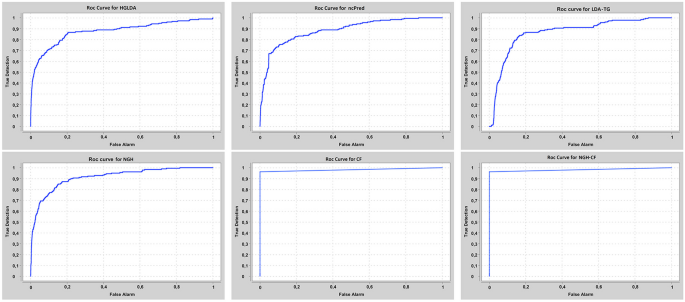
ROC curves for the compared methods on GS1
Another interesting issue is represented by the “agreement” between the different methods taking the same input, in terms of the returned best scoring LDAs. Table 2 shows the Jaccard Index computed between the proposed approaches and those receiving the same input, on the top \(5\%\) LDAs in the corresponding ranks, sorted from the best to the worst score values for each method. It emerges that results by HGLDA and ncPred have a small match with the other approaches (at most 0.23), while NGH-CF has high agreement with CF (0.74), as well as with NGH and LDA-TG (both 0.70). LDA-TG and CF present a sufficient match in their best predictions (0.59). This latter comparison based on agreement shows that approaches based on neighborhood analysis share a larger set of LDAs, in the top part of their ranks.
The proposed approaches have been compared also against other two recent methods from the literature, i.e., SIMCLDA and HGNNLDA, which receive in input different data than ours, including mRNA and drugs. For this reason, the more restrictive gold standard GS2 has been exploited for the comparison, where only lncRNAs and diseases having some correspondences with the additional input data of SIMCLDA and HGNNLDA are included. Figure 4 shows the comparison of the scored AUC on GS2, while Fig. 5 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change significantly on this other gold standard, moreover all the other approaches overcome SIMCLDA. On the other hand, HGNNLDA has a better performance than HGLDA, NcPred and LDA-TG, although it has a worse accuracy than NGH, CF and NGH-CF. The former confirms its superiority with regards to all considered approaches.
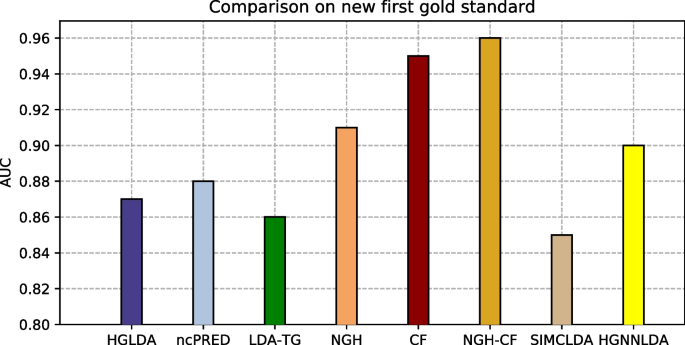
Comparison of the scored AUC on GS2
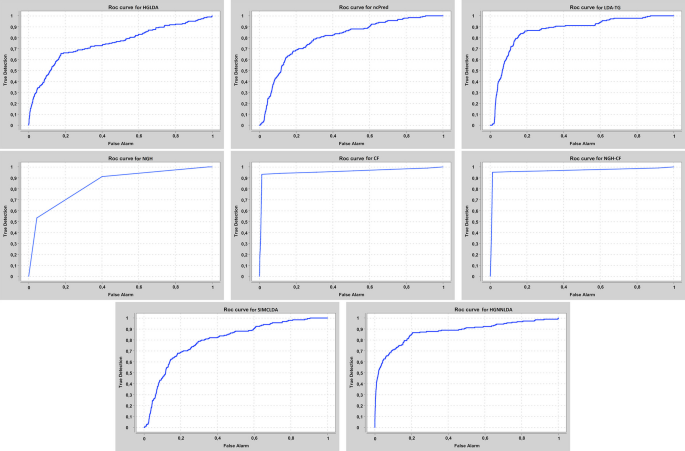
ROC curves for the compared methods on GS2
The proposed approaches have been compared also against LDAP-WMPS on GS3. Figure 6 shows the AUC values scored by all compared approaches on GS3, while Fig. 7 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change on this other gold standard, and LDAP-WMPS has better performance than the other approaches except for NGH, CF, NGH-CF and HGNNLDA.
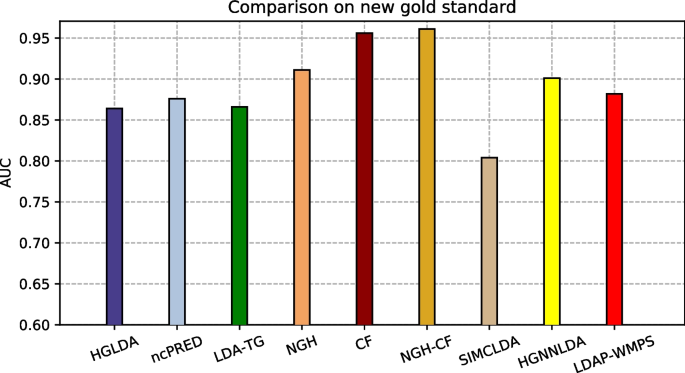
Comparison of the scored AUC on GS3
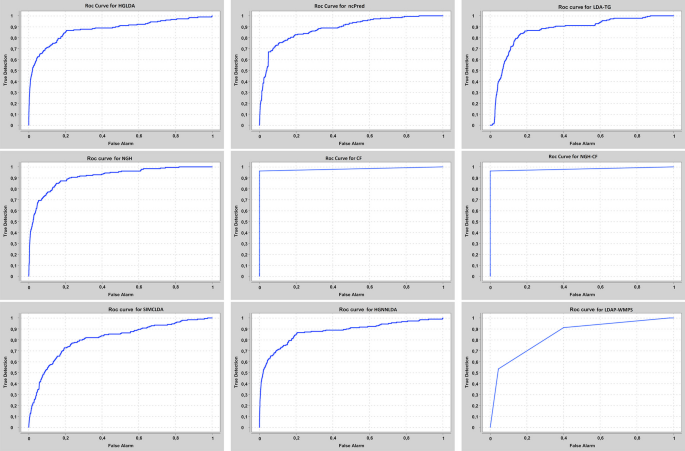
ROC curves for the compared methods on GS3
The AUPR values scored by the compared methods on GS1, GS2, and GS3 are shown in Fig. 8 , while the corresponding PR-curves are plotted in Fig. 9 . In particular, for GS1 results are analogous to the ROC analysis, with NGH-CF the best performing one, followed by CF and NGH, while HGLDA is the worst. On GS2, NGH-CF and CF keep their superiority, followed by SMCLDA and NGH, while HGLDA is yet the worst one. On GS3, NGH-CF is the first, Cf the second and both HGNNLDA and LDAP-WMPS outperform NGH, while HGLDA in this case slightly outperforms LDA-TG, ncPred and SMCLDA, which results to be the worst one.
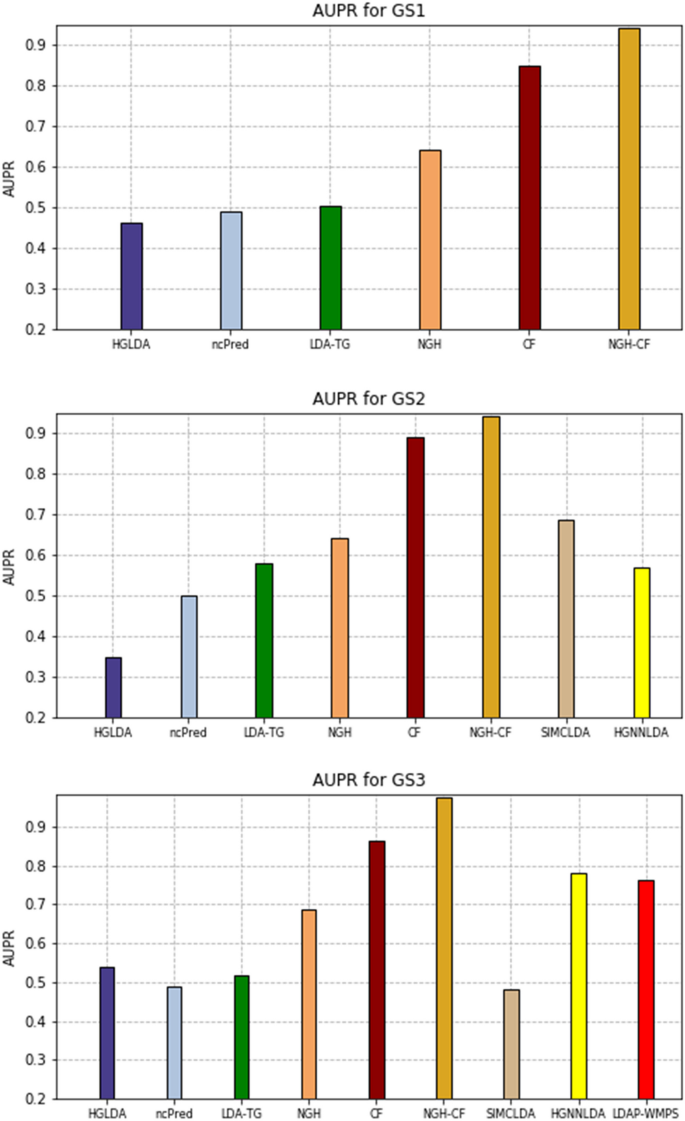
AUPR hystogram for the compared methods on GS1, GS2, GS3
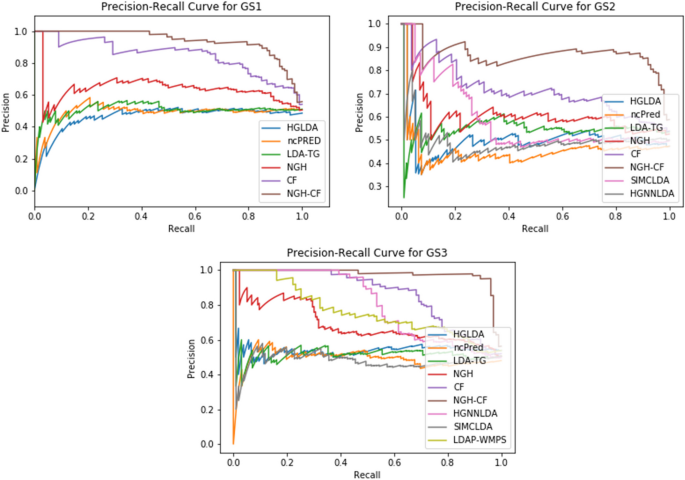
Precision-recall curves for the compared methods on GS1,GS2,GS3
Figures 10 , 11 and 12 show the F1-score values obtained, for all methods compared on GS1, GS2 and GS3, respectively, at the varying of a threshold fixed on the method score. In Tables 3 , 4 and 5 it is shown, for each gold standard, the highest value of F1-score obtained by each considered method, as well as the corresponding Precision and Recall values, and the minimum threshold value for which the highest F1-score value has been reached. On GS1 and GS2, the three best performing approaches are NGH-CF, CF and NGH, in this order. On GS3 the order is the same, and LDAP-WMPS performs equally to NGH.
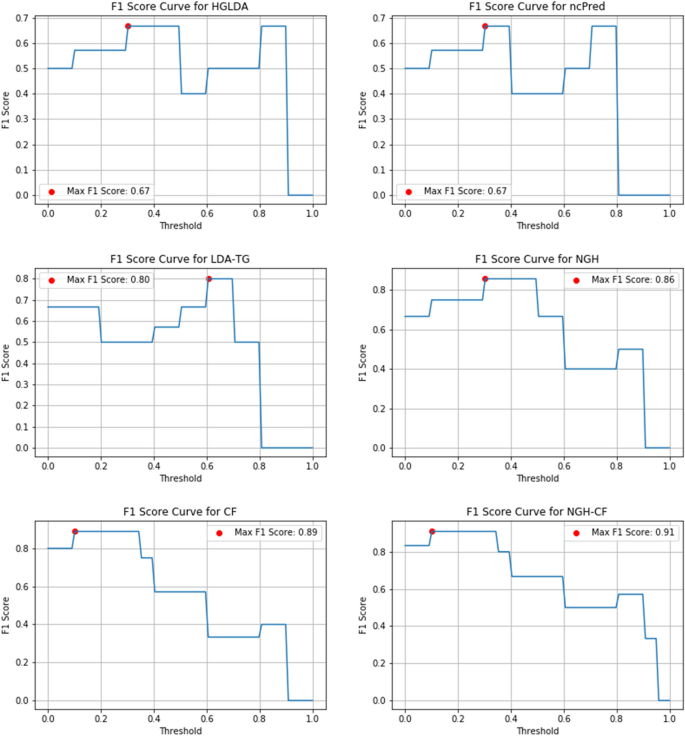
F1-score for the compared methods on GS1
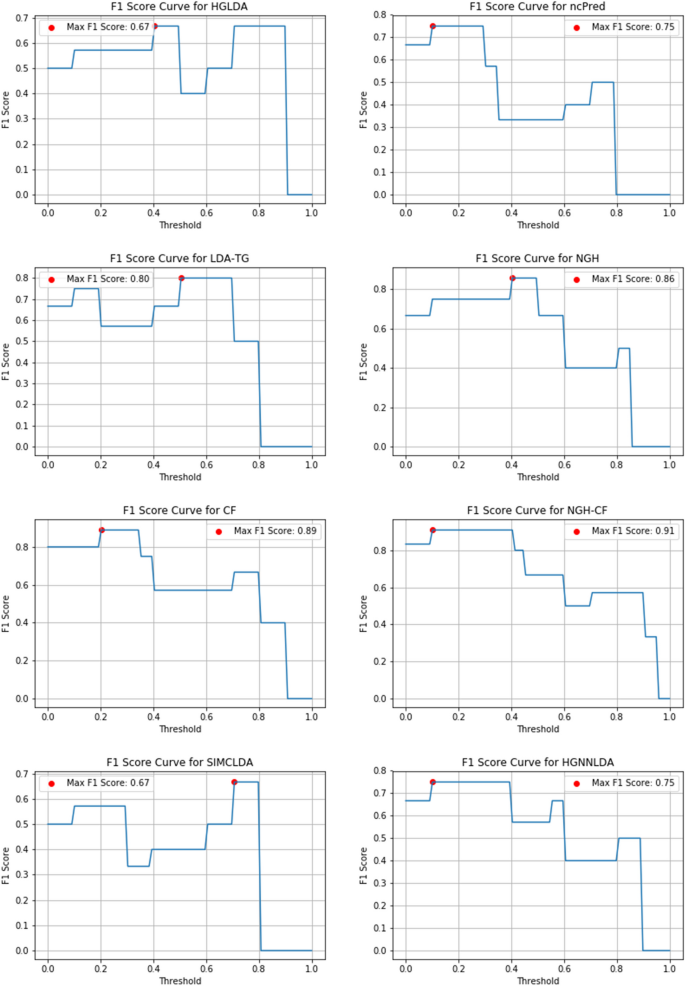
F1-Score for the compared methods on GS2
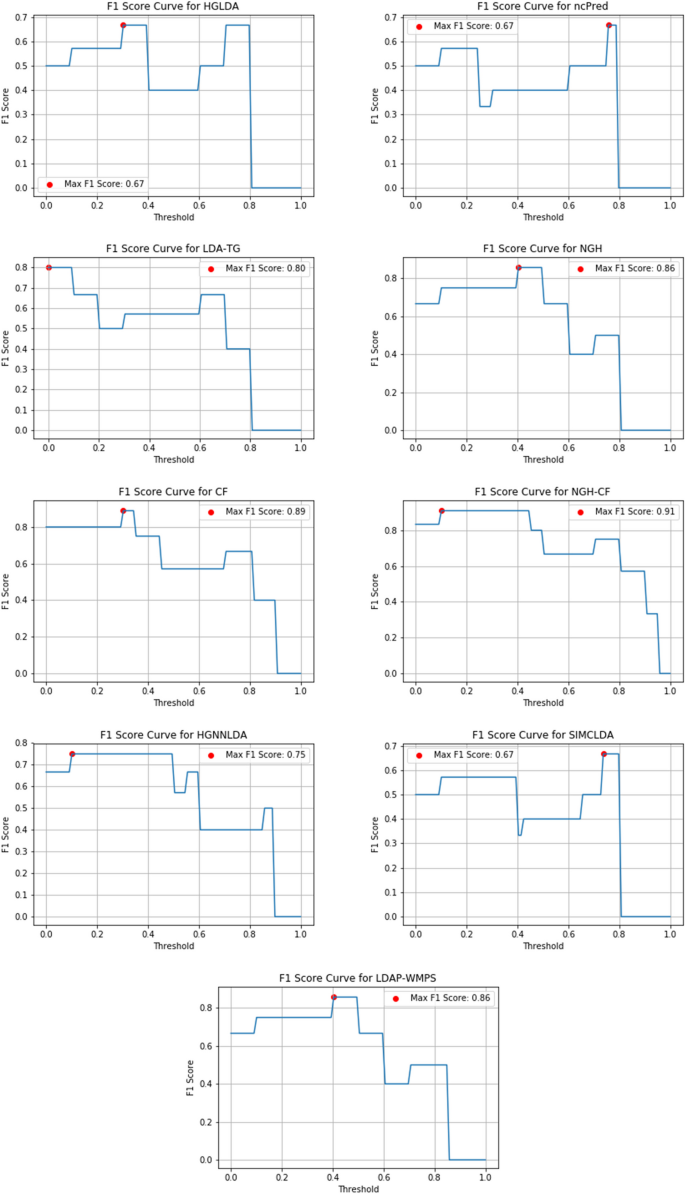
F1-Score for the compared methods on GS3
Robustness analysis
The main aim of the analysis discussed here is to measure to what extent the proposed methods are able to correctly recognize verified LDAs, even if part of the existing associations are missed, i.e., the sets of known and verified lncRNA-miRNA interactions and miRNA-disease associations are not complete. This is important to verify that the proposed approaches can provide reliable predictions also in presence of data incompleteness, that is often the case when lncRNAs are involved. Therefore, the robustness of each proposed method has been evaluated by performing progressive alterations of the input associations coming from the real datasets, according to the following three different criteria.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of lncRNA-miRNA interactions from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of miRNA-disease associations from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of both lncRNA-miRNA interactions and miRNA-disease associations (half and half), from the input data.
Tests summarized above have been performed for 20 times each. Tables 6 , 7 and 8 show the mean of the AUC values for NGH, CF and NGH-CF, respectively, over the 20 tests. In particular, all methods perform well on the three test typologies at \(5\%\) , the worst being NGH-CF, which however presents an average AUC equal to 0.84 for case 1), that is still a high value. NGH-CF is also the method that presents the best robustness on case 3), keeping the value of 0.92 also at \(20\%\) , while CF is the worst performing in case 3), indeed its average AUC decreases from 0.95 at \(5\%\) to 0.63 already at \(10\%\) , and then to 0.50 at \(20\%\) . This behaviour in case 3), where both lncRNA-miRNA interactions and miRNA-disease associations are progressively eliminated, deserves some observations. Indeed, results show that the combination of neighborhood analysis and collaborative filtering is the most robust one with regards to this perturbation, while collaborative filtering alone is the worst performing. On the other hand, CF results to be the most robust in case 1), where only lncRNA-miRNA interactions are eliminated, and this is due to the fact that CF does not take into account how many miRNAs are shared by pairs of lncRNAs. As for case 2), performance of all methods is comparable and generally good, possibly in consideration of the fact that a large number of miRNA-disease associations are available, therefore discarding small percentages of them does not affect largely the final prediction.
Comparison on specific situations
In this section further experimental tests are described, showing how well the considered methods perform in detecting specific situations, depicted through the synthetic dataset first, and then searched for in the real data. In particular, the basic observation here is that prediction approaches from the literature usually fail in detecting true LDAs, when the involved lncRNAs and diseases do not have a large number of shared miRNAs (referring to those approaches taking the same input than ours). The novel approaches we propose are particularly effective in managing the situation depicted above, through neighborhood analysis and collaborative filtering, allowing to detect similar behaviours shared by different lncRNAs, depending on the miRNAs they interact with.
For each set of LDAs defined in the synthetic data (i.e., set 1, set 2, and set 3), and for each tested method (i.e., HGLDA, NCPRED, NHG, CF, NGH-CF), Table 9 shows the percentage of LDAs in that set which is recognized at the top \(10\%\) , \(20\%\) , \(30\%\) , \(50\%\) of the rank of all LDAs, sorted by the score returned by the considered method. As an example, for HGLDA the \(32\%\) of LDAs of set 1 are located in the top \(10\%\) of its rank, where instead none LDAs in set 2 or 3 find place.
Looking at these results some interesting considerations come out. First of all, for the methods HGLDA, NCPRED, NHG and CF most associations of the set 1 are located in the top \(50\%\) of their corresponding ranks, while NGH-CF has a different behaviour. Indeed, it locates a lower number of such LDAs in the highest part of its rank than the other approaches, possibly due to the fact that it leaves room for a larger number of associations in the other two sets in the top ranked positions. As for LDAs in the set 2, all methods recognize some of them already in the top \(10\%\) , except for HGLDA, as alredy highlighted. The approaches able to recognize the larger percentages of these associations at the top \(50\%\) of their rank are NGH and NGH-CF. LDAs in the set 3 are the most difficult to recognize, due to the fact that the lncRNA and the disease do not share any miRNA in common. Indeed, the worst performing methods in this case are HGLDA, which is able to locate some of these associations only at the top \(50\%\) (according to the percentages we considered here), and NCPRED, which performs slightly better although it reaches the same percentage of located associations than HGLDA at \(50\%\) (the \(28\%\) ). As expected, approaches based on neighborhood analysis and collaborative filtering perform better, with the best one resulting to be NGH-CF.
In the previous section we have shown that all methods proposed here are able to detect specific situations, characterized by the fact that a lncRNA may have very few (or none) common miRNAs with a disease, and its neighbors share instead a large set of miRNAs with that disease. We have checked if this case occurs among the verified LDAs that our approaches find and their competitors do not. Table 10 shows, only by meaning of example, 10 experimentally verified LDAs, included in GS1, that are top ranked for the novel approaches proposed here, whereas they are in the bottom rank of the other approaches from the literature compared on GS1. Six out of such LDAs do not present any common miRNAs between the lncRNA and the disease, while four share only one miRNA. All involved lncRNAs present neighbors with a large number of miRNAs in common with the disease in that LDA, in accordance with the hypothesis that the ability in capturing this situation allows to obtain a better accuracy.
Survival analysis has been also performed by one of the TCGA Computational Tools, that is, TANRIC [ 41 ], on four of the pairs in Table 10 . In particular, those lncRNAs and diseases available in TANRIC have been chosen. Results are reported in Figures 13 , 14 , 15 and 16 , showing that the over-expression of the considered lncRNA determines a lower survival probability over the time, for all four considered cases.
Survival analysis related to SNHG16 and bladder neoplasm
Survival analysis related to CBR3-AS1 and prostate neoplasm
Survival analysis related to MALAT1 and bladder neoplasm
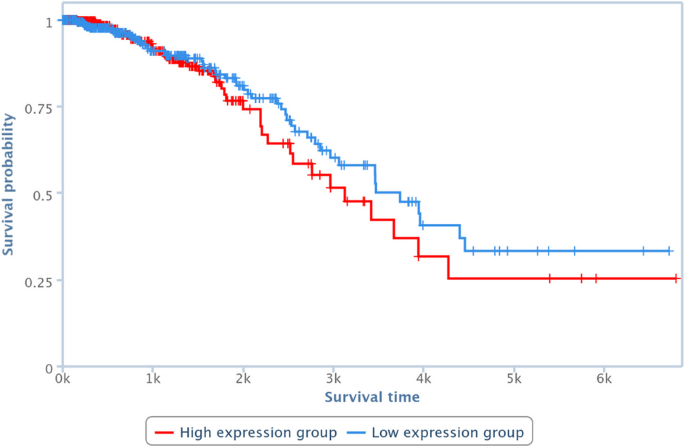
Survival analysis related to MEG3 and breast neoplasm
In the previous sections the effectiveness and robustness of the proposed approaches have been illustrated, showing that all three are able to return reliable predictions, as well as to detect specific situations which may occur in true predictions and are missed by competitors. Here we provide a discussion on some novel LDAs predicted by NGH, CF and NGH-CF.
Table 11 shows seven LDAs which are not present in the considered gold standards, and that have been returned by all three methods proposed here, with highest score. The first of these associations is between CDKN2B-AS1 and LEUKEMIA, confirmed by recent literature [ 42 , 43 ]. Indeed, CDKN2B-AS1 was found to be highly expressed in pediatric T-ALL peripheral blood mononuclear cells [ 42 ], moreover genome-wide association studies show that it is associated to Chronic Lymphocytic Leukaemia risk in Europeans [ 43 ]. As for the second association between DLEU2 and LEUKEMIA, DLEU2 is a long non-coding transcript with several splice variants, which has been identified by [ 44 ] through a comprehensive sequencing of a commonly deleted region in leukemia (i.e., the 13q14 region). Different investigations reported up regulation of this lncRNA in several types of cancers. The lncRNA H19 regulates GLIOMA angiogenesis [ 45 , 46 ], while MAP3K14 is one of the well-recognized biomarkers in the prognosis of renal cancer, which is reminiscent of the pancreatic metastasis from renal cell carcinoma [ 47 ]. MEG3 has been recently found to be important for the prediction of LEUKEMIA risk [ 48 ]. Multiple studies have shown that MIR155HG is highly expressed in diffuse large B-cell (DLBC) lymphoma and primary mediastinal B-cell lymphoma, and in chronic lymphocytic leukemia. The transcription factor MYB activates MIR155HG activity, which causes the epigenetic state of MIR155HG to be dysregulated and causes an abnormal increase in MIR155 [ 49 ]. Also the last top-ranked association in Table 11 between TUG1 and NON-SMALL CELL LUNG CARCINOMA has found evidence in the literature [ 50 , 51 , 52 ].
Tables 12 , 13 , and 14 show the top 100 (sorted by the scores returned by each method) novel LDA predictions that NGH and CF, NGH and NGH-CF, CF and NGH-CF have in common, respectively. Many of the lncRNAs involved in such top-ranked LDAs are not yet characterized in the literature, therefore results presented here may be considered a first attempt to provide novel knowledge about them, through their inferred association with known diseases.
We have explored the application of neighborhood analysis, combined with collaborative filtering, for the improvement of LDAs prediction accuracy. The three approaches proposed here have been evaluated and compared first against their direct competitors from the literature, i.e., the other methods which also use lncRNA-miRNA interactions and miRNA-disease associations, without exploiting a priori known LDAs. It results that all methods proposed here are able to outperform direct competitors, the best one (NGH-CF) also significantly (AUC equal to 0.966 against the 0.886 by NCPRED). In particular, it has been shown that the improvement in accuracy is due to the fact that our approaches capture specific situations neglected by competitors, relying on similar lncRNAs behaviour in terms of their interactions with the considered intermediate molecules (i.e., miRNAs). The proposed approaches have been then compared also against other recent methods, taking different inputs (e.g., integrative approaches), and the experimental evaluation shows that they are able to outperform them as well.
It is worth pointing out the importance of providing reliable data in input to the LDAs prediction approaches. As discussed in this manuscript, information on the lncRNAs relationships with other molecules, and between intermediate molecules and diseases, is provided in input to the proposed approaches. Reliable datasets have been used to perform the experimental analysis provided here. However, as the user may provide also different input datasets, it is important to point out that the reliability of the obtained predictions strictly depends on that of input information.
As neighborhood analysis has resulted to be effective in characterizing lncRNAs with regards to their association with known diseases, we plan to apply it also for predicting possible common functions among lncRNAs, for example by clustering them according to their interactions, which has shown to be successful for other types of molecules [ 53 ]. Moreover, due to the success of integrative approaches on the analysis of biological data [ 54 ], we expect that including other types of intermediate molecules, such as for example genes and proteins, in the main pipeline of the proposed approaches may further improve their accuracy.
In conclusion, the use of reliable input data and the integration of different types of information coming from molecular interactions seem to be the most promising future directions for LDAs prediction.
Availability of data and materials
The source code is available at: https://github.com/marybonomo/LDAsPredictionApproaches.git In particular, executable software for NGH, CF, and NGH-CF are provided, as well as syntetic and real input datasets used here; the three different gold standard datasets GS1, GS2, GS3; the final obtained results.
Medico-Salsench E, et al. The non-coding genome in genetic brain disorders: New targets for therapy? Essays Biochem. 2021;65(4):671–83.
Article CAS PubMed PubMed Central Google Scholar
Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Article CAS PubMed Google Scholar
Zhao H, Shi J, Zhang Y, et al. LncTarD: a manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucl Acids Res. 2019;48(D1):D118–D126. ISSN: 0305-1048.
Liao Q, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co- expression network. Nuc Acids Res. 2011;39:3864–78.
Article CAS Google Scholar
Chen X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinf. 2017;18(4):558–76.
CAS Google Scholar
Wang B, et al. lncRNA-disease association prediction based on matrix decomposition of elastic network and collaborative filtering. Sci Rep. 2022;12:7.
Google Scholar
He J, et al. HOPEXGB: a consensual model for predicting miRNA/lncRNA-disease associations using a heterogeneous disease-miRNA-lncRNA information network. J Chem Inf Model 2023
Zhong H, et al. Association filtering and generative adversarial networks for predicting lncRNA-associated disease. BMC Bioinf. 2023;24(1):234.
Dengju Y, et al. GCNFORMER: graph convolutional network and transformer for predicting lncRNA-disease associations. BMC Bioinf. 2024;25(1):5.
Article Google Scholar
Alaimo S, Giugno R, Pulvirenti A. ncPred: ncRNA-disease association prediction through Tripartite network-based inference. Front Bioeng Biot. 2014;2:71.
Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5:13186.
Lu C, et al. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics. 2018;34(19):3357–64.
Xuan Z, Li J, Yu X, Feng J, et al. A probabilistic matrix factorization method for identifying lncRNA-disease associations. Genes 2019;10(2)
Du X, et al. lncRNA-disease association prediction method based on the nearest neighbor matrix completion model. Sci Rep. 2022;12(1):21653.
Wang L, et al. Prediction of lncRNA-disease association based on a Laplace normalized random walk with restart algorithm on heterogeneous networks. BMC Bioinf. 2022;23(1):1–20.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: taxonomy, trends and challenges of computational models. Brief Bioinf. 2022;23(5):bbac358.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: experimental results, databases, webservers and data fusion. Brief Bioinf. 2022;23(6):bbac397.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: towards systematic evaluation of computational models. Brief Bioinf. 2022;23(6):bbac407.
Chen X, Yan G. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29(20):2617–24.
Xie G, et al. SKF-LDA: similarity kernel fusion for predicting lncRNA-disease association. Mol Therapy-Nucleic Acids. 2019;18:45–55.
Liu D, et al. HGNNLDA: predicting lncRNA-drug sensitivity associations via a dual channel hypergraph neural network. IEEE/ACM transactions on computational biology and bioinformatics, 2023;1–11.
Zhang Y, et al. LDAI-ISPS: lncRNA-disease associations inference based on integrated space projection scores. Int J Molecular Sci. 2020;21(4):1508.
Liang Y, et al. MAGCNSE: predicting lncRNA-disease associations using multi-view attention graph convolutional network and stacking ensemble model. BMC Bioinf. 2022;23(1):189.
Bonomo M, La Placa A, Rombo SE. Prediction of lncRNA-disease associations from tripartite graphs. In: Heterogeneous data management, polystores, and analytics for healthcare - VLDB workshops, poly 2020 and DMAH 2020, virtual event, August 31 and September 4, 2020, Revised Selected Papers. Springer, Berlin, 2020;205–210. ISSN: 978-3-030-71054-5
Xie G, et al. Predicting lncRNA-disease associations based on combining selective similarity matrix fusion and bidirectional linear neighborhood label propagation. Brief Bioinform. 2023;24(1):bbac595.
Article PubMed Google Scholar
Cheng L, et al. ntNetLncSim: an integrative network analysis method to infer human lncRNA functional similarity. Oncotarget. 2016;7(30):47864–74.
Article PubMed PubMed Central Google Scholar
Guangyuan F, et al. Matrix factorization-based data fusion for the prediction of lncRNA-disease associations. Bioinformatics. 2018;34:1529–37.
Xie G, et al. RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol Genet Genom. 2021;296:473–83.
Wang B, et al. lncRNA-disease association prediction based on the weight matrix and projection score. PLOS One. 2023;18(1): e0278817.
Duan R, Jiang C, Jain HK. Combining review-based collaborative filtering and matrix factorization: a solution to rating’s sparsity problem”. Decis Support Syst 2022;156:113748. ISSN: 0167–9236.
Koren Y, Bell R, Volinsky C. Matrix factorization techniques for recommender systems. Computer. 2009;42(8):30–7.
Parida L, Pizzi C, Rombo SE. Irredundant tandem motifs. Theoret Comput Sci. 2014;525:89–102.
Bonomo M, et al. Topological ranks reveal functional knowledge encoded in biological networks: a comparative analysis. Brief Bioinform. 2022;23(3):bbac101.
Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–74.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLOS One. 2015;10(3): e0118432.
Li J, et al. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013;42:D92–7.
Li Y, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–4.
Chen G, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6.
Gao Y, et al. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021;49(D1):D1251–8.
Cui T, et al. MNDR v2. 0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46(D1):D371–4.
CAS PubMed Google Scholar
Li J, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–37.
Chen L, et al. lncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335-3p/TRAF5 axis. In: Anti-cancer drugs, Wolters Kluwer Health, Inc. (2020)
Song C, et al. CDKN2B-AS1: an indispensable long non-coding RNA in multiple diseases. Current Pharm Des. 2020;26(41):5335–46.
Ghafouri-Fard S, et al. Deleted in lymphocytic leukemia 2 (DLEU2): an lncRNA with dissimilar roles in different cancers. Biomed Pharmacother. 2021;133: 111093.
Jia P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–69.
Liu Z, et al. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1 α /VEGFaxis. Neoplasma. 2020;67(1):111–8.
Zhou S, et al. A novel immune-related gene prognostic Index (IRGPI) in pancreatic adenocarcinoma (PAAD) and its implications in the tumor microenvironment. Cancers. 2022;14(22):5652.
Pei J, et al. Novel contribution of long non-coding RNA MEG3 genotype to prediction of childhood leukemia risk. Cancer Genom Proteom. 2022;19(1):27–34.
Peng L, et al. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8(17):7161–73.
Zhang E, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5(5):e1243–e1243.
Lin P, et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:1–10.
Niu Y, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):1–13.
Pizzuti C, Rombo SE. An evolutionary restricted neighborhood search clustering approach for PPI networks. Neurocomputing. 2014;145:53–61.
Rombo SE, Ursino D (2021) Integrative bioinformatics and omics data source interoperability in the next-generation sequencing era
Download references
Acknowledgements
The authors are grateful to the Anonymous Reviewers, for the constructive and useful suggestions that allowed to significantly improve the quality of this manuscript. Some of the results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga .
PRIN “multicriteria data structures and algorithms: from compressed to learned indexes, and beyond”, Grant No. 2017WR7SHH, funded by MIUR (closed). “Modelling and analysis of big knowledge graphs for web and medical problem solving” (CUP: E55F22000270001), “Computational Approaches for Decision Support in Precision Medicine” (CUP:E53C22001930001), and “Knowledge graphs e altre rappresentazioni compatte della conoscenza per l’analisi di big data” (CUP: E53C23001670001), funded by INdAM GNCS 2022, 2023, 2024 projects, respectively. “Models and Algorithms relying on knowledge Graphs for sustainable Development goals monitoring and Accomplishment - MAGDA” (CUP: B77G24000050001), funded by the European Union under the PNRR program related to “Future Artificial Intelligence - FAIR”.
Author information
Authors and affiliations.
Kazaam Lab s.r.l., Palermo, Italy
Mariella Bonomo & Simona E. Rombo
Department of Mathematics and Computer Science, University of Palermo, Palermo, Italy
Simona E. Rombo
You can also search for this author in PubMed Google Scholar
Contributions
MB and SER equally contributed to the research presented in this manuscript. MB implemented and run the software, SER performed the analysis of results. Both authors wrote and reviewed the entire manuscript.
Corresponding author
Correspondence to Mariella Bonomo .
Ethics declarations
Ethics approval and consent to participate.
Not Applicable
Consent for publication
Competing interests.
SER is editor of BMC Bionformatics. MB has no Conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Bonomo, M., Rombo, S.E. Neighborhood based computational approaches for the prediction of lncRNA-disease associations. BMC Bioinformatics 25 , 187 (2024). https://doi.org/10.1186/s12859-024-05777-8
Download citation
Received : 13 December 2023
Accepted : 11 April 2024
Published : 13 May 2024
DOI : https://doi.org/10.1186/s12859-024-05777-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- LncRNA-disease associations
- Molecular interactions
- Bioinformatics
- Long non-coding RNA
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
Analytical Methods
Application of surfactants in the electrochemical and biosensing of biomolecules and drug molecules.
Realizing sensitive and efficient detection of biomolecules and drug molecules is of great significance. Among the detection methods that have been proposed, electrochemical sensing is favored for its outstanding advantages such as simple operation, low cost, fast response and high sensitivity. The unique structure and properties of surfactants have led to a wide range of applications in the field of electrochemical sensors and biosensors for biomolecules and drug molecules. Through the comparative analysis of reported works, this paper summarizes the application modes of surfactants in electrochemical sensors and biosensors for biomolecules and drug molecules, explores the possible electrocatalytic mechanism of their action, and looks forward to the development trend of their applications. This review is expected to provide some new ideas for subsequent related research work.
- This article is part of the themed collection: Analytical Methods Review Articles 2024
Article information
Download citation, permissions.
T. Chen, S. Zhang, C. Zhu, C. Liu, X. Liu, S. S. Hu, D. Zheng and J. Zhang, Anal. Methods , 2024, Accepted Manuscript , DOI: 10.1039/D4AY00313F
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page .
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page .
Read more about how to correctly acknowledge RSC content .
Social activity
Search articles by author.
This article has not yet been cited.
Advertisements
The Federal Register
The daily journal of the united states government, request access.
Due to aggressive automated scraping of FederalRegister.gov and eCFR.gov, programmatic access to these sites is limited to access to our extensive developer APIs.
If you are human user receiving this message, we can add your IP address to a set of IPs that can access FederalRegister.gov & eCFR.gov; complete the CAPTCHA (bot test) below and click "Request Access". This process will be necessary for each IP address you wish to access the site from, requests are valid for approximately one quarter (three months) after which the process may need to be repeated.
An official website of the United States government.
If you want to request a wider IP range, first request access for your current IP, and then use the "Site Feedback" button found in the lower left-hand side to make the request.
- Open access
- Published: 13 May 2024
SCIPAC: quantitative estimation of cell-phenotype associations
- Dailin Gan 1 ,
- Yini Zhu 2 ,
- Xin Lu 2 , 3 &
- Jun Li ORCID: orcid.org/0000-0003-4353-5761 1
Genome Biology volume 25 , Article number: 119 ( 2024 ) Cite this article
175 Accesses
2 Altmetric
Metrics details
Numerous algorithms have been proposed to identify cell types in single-cell RNA sequencing data, yet a fundamental problem remains: determining associations between cells and phenotypes such as cancer. We develop SCIPAC, the first algorithm that quantitatively estimates the association between each cell in single-cell data and a phenotype. SCIPAC also provides a p -value for each association and applies to data with virtually any type of phenotype. We demonstrate SCIPAC’s accuracy in simulated data. On four real cancerous or noncancerous datasets, insights from SCIPAC help interpret the data and generate new hypotheses. SCIPAC requires minimum tuning and is computationally very fast.
Single-cell RNA sequencing (scRNA-seq) technologies are revolutionizing biomedical research by providing comprehensive characterizations of diverse cell populations in heterogeneous tissues [ 1 , 2 ]. Unlike bulk RNA sequencing (RNA-seq), which measures the average expression profile of the whole tissue, scRNA-seq gives the expression profiles of thousands of individual cells in the tissue [ 3 , 4 , 5 , 6 , 7 ]. Based on this rich data, cell types may be discovered/determined in an unsupervised (e.g., [ 8 , 9 ]), semi-supervised (e.g., [ 10 , 11 , 12 , 13 ]), or supervised manner (e.g., [ 14 , 15 , 16 ]). Despite the fast development, there are still limitations with scRNA-seq technologies. Notably, the cost for each scRNA-seq experiment is still high; as a result, most scRNA-seq data are from a single or a few biological samples/tissues. Very few datasets consist of large numbers of samples with different phenotypes, e.g., cancer vs. normal. This places great difficulties in determining how a cell type contributes to a phenotype based on single-cell studies (especially if the cell type is discovered in a completely unsupervised manner or if people have limited knowledge of this cell type). For example, without having single-cell data from multiple cancer patients and multiple normal controls, it could be hard to computationally infer whether a cell type may promote or inhibit cancer development. However, such association can be critical for cancer research [ 17 ], disease diagnosis [ 18 ], cell-type targeted therapy development [ 19 ], etc.
Fortunately, this difficulty may be overcome by borrowing information from bulk RNA-seq data. Over the past decade, a considerable amount of bulk RNA-seq data from a large number of samples with different phenotypes have been accumulated and made available through databases like The Cancer Genome Atlas (TCGA) [ 20 ] and cBioPortal [ 21 , 22 ]. Data in these databases often contain comprehensive patient phenotype information, such as cancer status, cancer stages, survival status and time, and tumor metastasis. Combining single-cell data from a single or a few individuals and bulk data from a relatively large number of individuals regarding a particular phenotype can be a cost-effective way to determine how a cell type contributes to the phenotype. A recent method Scissor [ 23 ] took an essential step in this direction. It uses single-cell and bulk RNA-seq data with phenotype information to classify the cells into three discrete categories: Scissor+, Scissor−, and null cells, corresponding to cells that are positively associated, negatively associated, and not associated with the phenotype.
Here, we present a method that takes another big step in this direction, which is called Single-Cell and bulk data-based Identifier for Phenotype Associated Cells or SCIPAC for short. SCIPAC enables quantitative estimation of the strength of association between each cell in a scRNA-seq data and a phenotype, with the help of bulk RNA-seq data with phenotype information. Moreover, SCIPAC also enables the estimation of the statistical significance of the association. That is, it gives a p -value for the association between each cell and the phenotype. Furthermore, SCIPAC enables the estimation of association between cells and an ordinal phenotype (e.g., different stages of cancer), which could be informative as people may not only be interested in the emergence/existence of cancer (cancer vs. healthy, a binary problem) but also in the progression of cancer (different stages of cancer, an ordinal problem).
To study the performance of SCIPAC, we first apply SCIPAC to simulated data under three schemes. SCIPAC shows high accuracy with low false positive rates. We further show the broad applicability of SCIPAC on real datasets across various diseases, including prostate cancer, breast cancer, lung cancer, and muscular dystrophy. The association inferred by SCIPAC is highly informative. In real datasets, some cell types have definite and well-studied functions, while others are less well-understood: their functions may be disease-dependent or tissue-dependent, and they may contain different sub-types with distinct functions. In the former case, SCIPAC’s results agree with current biological knowledge. In the latter case, SCIPAC’s discoveries inspire the generation of new hypotheses regarding the roles and functions of cells under different conditions.
An overview of the SCIPAC algorithm
SCIPAC is a computational method that identifies cells in single-cell data that are associated with a given phenotype. This phenotype can be binary (e.g., cancer vs. normal), ordinal (e.g., cancer stage), continuous (e.g., quantitative traits), or survival (i.e., survival time and status). SCIPAC uses input data consisting of three parts: single-cell RNA-seq data that measures the expression of p genes in m cells, bulk RNA-seq data that measures the expression of the same set of p genes in n samples/tissues, and the statuses/values of the phenotype of the n bulk samples/tissues. The output of SCIPAC is the strength and the p -value of the association between each cell and the phenotype.
SCIPAC proposes the following definition of “association” between a cell and a phenotype: A group of cells that are likely to play a similar role in the phenotype (such as cells of a specific cell type or sub-type, cells in a particular state, cells in a cluster, cells with similar expression profiles, or cells with similar functions) is considered to be positively/negatively associated with a phenotype if an increase in their proportion within the tissue likely indicates an increased/decreased probability of the phenotype’s presence. SCIPAC assigns the same association to all cells within such a group. Taking cancer as the phenotype as an example, if increasing the proportion of a cell type indicates a higher chance of having cancer (binary), having a higher cancer stage (ordinal), or a higher hazard rate (survival), all cells in this cell type is positively associated with cancer.
The algorithm of SCIPAC follows the following four steps. First, the cells in the single-cell data are grouped into clusters according to their expression profiles. The Louvain algorithm from the Seurat package [ 24 , 25 ] is used as the default clustering algorithm, but the user may choose any clustering algorithm they prefer. Or if information of the cell types or other groupings of cells is available a priori, it may be supplied to SCIPAC as the cell clusters, and this clustering step can be skipped. In the second step, a regression model is learned from bulk gene expression data with the phenotype. Depending on the type of the phenotype, this model can be logistic regression, ordinary linear regression, proportional odds model, or Cox proportional hazards model. To achieve a higher prediction power with less variance, by default, the elastic net (a blender of Lasso and ridge regression [ 26 ]) is used to fit the model. In the third step, SCIPAC computes the association strength \(\Lambda\) between each cell cluster and the phenotype based on a mathematical formula that we derive. Finally, the p -values are computed. The association strength and its p -value between a cell cluster and the phenotype are given to all cells in the cluster.
SCIPAC requires minimum tuning. When the cell-type information is given in step 1, SCIPAC does not have any (hyper)parameter. Otherwise, the Louvain algorithm used in step 1 has a “resolution” parameter that controls the number of cell clusters: a larger resolution results in more clusters. SCIPAC inherits this parameter as its only parameter. Since SCIPAC gives the same association strength and p -value to cells from the same cluster, this parameter also determines the resolution of results provided by SCIPAC. Thus, we still call it “resolution” in SCIPAC. Because of its meaning, we recommend setting it so that the number of cell clusters given by the clustering algorithm is comparable to, or reasonably larger than, the number of cell types (or sub-types) in the data. We will see that the performance of SCIPAC is insensitive to this resolution parameter, and the default value 2.0 typically works well.
The details of the SCIPAC algorithm are given in the “ Methods ” section.
Performance in simulated data
We assess the performance of SCIPAC in simulated data under three different schemes. The first scheme is simple and consists of only three cell types. The second scheme is more complicated and consists of seven cell types, which better imitates actual scRNA-seq data. In the third scheme, we simulate cells under different cell development stages to test the performance of SCIPAC under an ordinal phenotype. Details of the simulation are given in Additional file 1.
Simulation scheme I
Under this scheme, the single-cell data consists of three cell types: one is positively associated with the phenotype, one is negatively associated, and the third is not associated (we call it “null”). Figure 1 a gives the UMAP [ 27 ] plot of the three cell types, and Fig. 1 b gives the true associations of these three cell types with the phenotype, with red, blue, and light gray denoting positive, negative, and null associations.
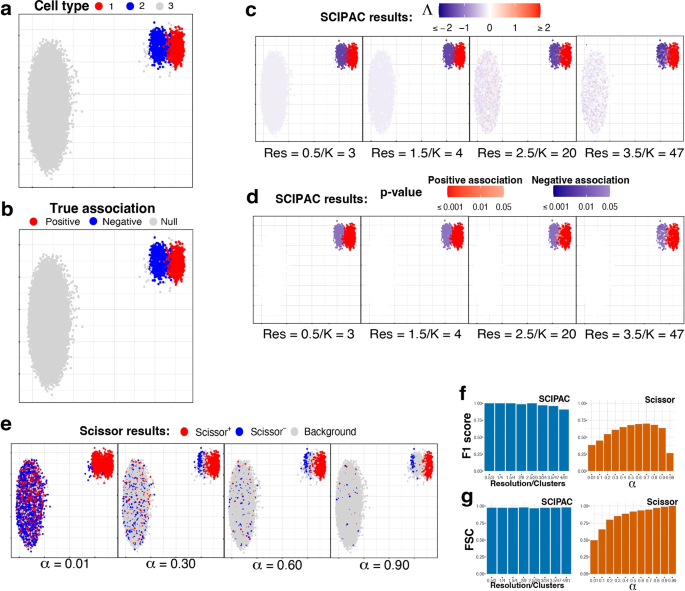
UMAP visualization and numeric measures of the simulated data under scheme I. All the plots in a–e are scatterplots of the two dimensional single-cell data given by UMAP. The x and y axes represent the two dimensions, and their scales are not shown as their specific values are not directly relevant. Points in the plots represents single cells, and they are colored differently in each subplot to reflect different information/results. a Cell types. b True associations. The association between cell types 1, 2, and 3 and the phenotype are positive, negative, and null, respectively. c Association strengths \(\Lambda\) given by SCIPAC under different resolutions. Red/blue represents the sign of \(\Lambda\) , and the shade gives the absolute value of \(\Lambda\) . Every cell is colored red or blue since no \(\Lambda\) is exactly zero. Below each subplot, Res stands for resolution, and K stands for the number of cell clusters given by this resolution. d p -values given by SCIPAC. Only cells with p -value \(< 0.05\) are colored red (positive association) or blue (negative association); others are colored white. e Results given by Scissor under different \(\alpha\) values. Red, blue, and light gray stands for Scissor+, Scissor−, and background (i.e., null) cells. f F1 scores and g FSC for SCIPAC and Scissor under different parameter values. For SCIPAC, each bar is the value under a resolution/number of clusters. For Scissor, each bar is the value under an \(\alpha\)
We apply SCIPAC to the simulated data. For the resolution parameter (see the “ Methods ” section), values 0.5, 1.0, and 1.5 give 3, 4, and 4 clusters, respectively, close to the actual number of cell types. They are good choices based on the guidance for choosing this parameter. To show how SCIPAC behaves under parameter misspecification, we also set the resolution up to 4.0, which gives a whopping 61 clusters. Figure 1 c and d give the association strengths \(\Lambda\) and the p -values given by four different resolutions (results under other resolutions are provided in Additional file 1: Fig. S1 and S2). In Fig. 1 c, red and blue denote positive and negative associations, respectively, and the shade of the color represents the strength of the association, i.e., the absolute value of \(\Lambda\) . Every cell is colored blue or red, as none of \(\Lambda\) is exactly zero. In Fig. 1 d, red and blue denote positive and negative associations that are statistically significant ( p -value \(< 0.05\) ). Cells whose associations are not statistically significant ( p -value \(\ge 0.05\) ) are shown in white. To avoid confusion, it is worth repeating that cells that are colored in red/blue in Fig. 1 c are shown in red/blue in Fig. 1 d only if they are statistically significant; otherwise, they are colored white in Fig. 1 d.
From Fig. 1 c, d (as well as Additional file 1: Fig. S1 and S2), it is clear that the results of SCIPAC are highly consistent under different resolution values, including both the estimated association strengths and the p -values. It is also clear that SCIPAC is highly accurate: most truly associated cells are identified as significant, and most, if not all, truly null cells are identified as null.
As the first algorithm that quantitatively estimates the association strength and the first algorithm that gives the p -value of the association, SCIPAC does not have a real competitor. A previous algorithm, Scissor, is able to classify cells into three discrete categories according to their associations with the phenotype. So, we compare SCIPAC with Scissor in respect of the ability to differentiate positively associated, negatively associated, and null cells.
Running Scissor requires tuning a parameter called \(\alpha\) , which is a number between 0 and 1 that balances the amount of regularization for the smoothness and for the sparsity of the associations. The Scissor R package does not provide a default value for this \(\alpha\) or a function to help select this value. The Scissor paper suggests the following criterion: “the number of Scissor-selected cells should not exceed a certain percentage of total cells (default 20%) in the single-cell data. In each experiment, a search on the above searching list is performed from the smallest to the largest until a value of \(\alpha\) meets the above criteria.” In practice, we have found that this criterion does not often work properly, as the truly associated cells may not compose 20% of all cells in actual data. Therefore, instead of setting \(\alpha\) to any particular value, we set \(\alpha\) values that span the whole range of \(\alpha\) to see the best possible performance of Scissor.
The performance of Scissor in our simulation data under four different \(\alpha\) values are shown in Fig. 1 e, and results under more \(\alpha\) values are shown in Additional file 1: Fig. S3. In the figures, red, blue, and light gray denote Scissor+, Scissor−, and null (called “background” in Scissor) cells, respectively. The results of Scissor have several characteristics different from SCIPAC. First, Scissor does not give the strength or statistical significance of the association, and thus the colors of the cells in the figures do not have different shades. Second, different \(\alpha\) values give very different results. Greater \(\alpha\) values generally give fewer Scissor+ and Scissor− cells, but there are additional complexities. One complexity is that the Scissor+ (or Scissor−) cells under a greater \(\alpha\) value are not a strict subset of Scissor+ (or Scissor−) cells under a smaller \(\alpha\) value. For example, the number of truly negatively associated cells detected as Scissor− increases when \(\alpha\) increases from 0.01 to 0.30. Another complexity is that the direction of the association may flip as \(\alpha\) increases. For example, most cells of cell type 2 are identified as Scissor+ under \(\alpha =0.01\) , but many of them are identified as Scissor− under larger \(\alpha\) values. Third, Scissor does not achieve high power and low false-positive rate at the same time under any \(\alpha\) . No matter what the \(\alpha\) value is, there is only a small proportion of cells from cell type 2 that are correctly identified as negatively associated, and there is always a non-negligible proportion of null cells (i.e., cells from cell type 3) that are incorrectly identified as positively or negatively associated. Fourth, Scissor+ and Scissor− cells can be close to each other in the figure, even under a large \(\alpha\) value. This means that cells with nearly identical expression profiles are detected to be associated with the phenotype in opposite directions, which can place difficulties in interpreting the results.
SCIPAC overcomes the difficulties of Scissor and gives results that are more informative (quantitative strengths with p -values), more accurate (both high power and low false-positive rate), less sensitive to the tuning parameter, and easier to interpret (cells with similar expression typically have similar associations to the phenotype).
SCIPAC’s higher accuracy in differentiating positively associated, negatively associated, and null cells than Scissors can also be measured numerically using the F1 score and the fraction of sign correctness (FSC). F1, which is the harmonic mean of precision and recall, is a commonly used measure of calling accuracy. Note that precision and recall are only defined for two-class problems, which try to classify desired signals/discoveries (so-called “positives”) against noises/trivial results (so-called “negatives”). Our case, on the other hand, is a three-class problem: positive association, negative association, and null. To compute F1, we combine the positive and negative associations and treat them as “positives,” and treat null as “negatives.” This F1 score ignores the direction of the association; thus, it alone is not enough to describe the performance of an association-detection algorithm. For example, an algorithm may have a perfect F1 score even if it incorrectly calls all negative associations positive. To measure an algorithm’s ability to determine the direction of the association, we propose a statistic called FSC, defined as the fraction of true discoveries that also have the correct direction of the association. The F1 score and FSC are numbers between 0 and 1, and higher values are preferred. A mathematical definition of these two measures is given in Additional file 1.
Figure 1 f, g show the F1 score and FSC of SCIPAC and Scissor under different values of tuning parameters. The F1 score of Scissor is between 0.2 and 0.7 under different \(\alpha\) ’s. The FSC of Scissor increases from around 0.5 to nearly 1 as \(\alpha\) increases, but Scissor does not achieve high F1 and FSC scores at the same time under any \(\alpha\) . On the other hand, the F1 score of SCIPAC is close to perfection when the resolution parameter is properly set, and it is still above 0.90 even if the resolution parameter is set too large. The FSC of SCIPAC is always above 0.96 under different resolutions. That is, SCIPAC achieves high F1 and FSC scores simultaneously under a wide range of resolutions, representing a much higher accuracy than Scissor.
Simulation scheme II
This more complicated simulation scheme has seven cell types, which are shown in Fig. 2 a. As shown in Fig. 2 b, cell types 1 and 3 are negatively associated (colored blue), 2 and 4 are positively associated (colored red), and 5, 6, and 7 are not associated (colored light gray).
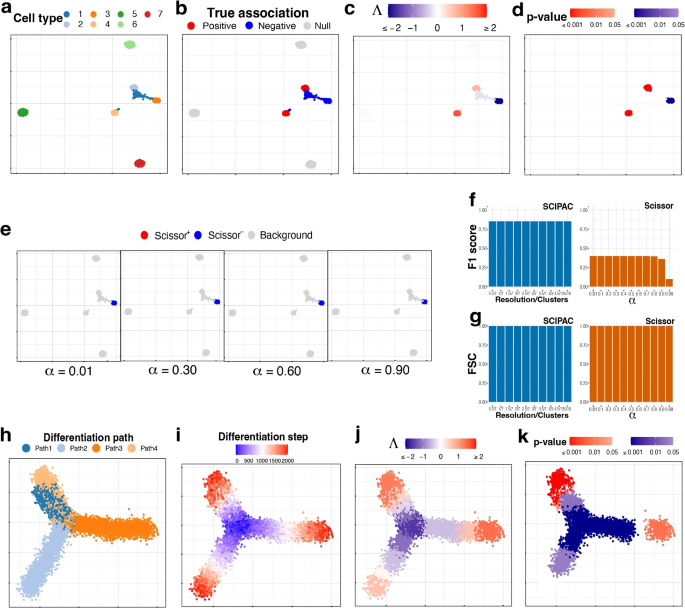
UMAP visualization of the simulated data under a–g scheme II and h–k scheme III. a Cell types. b True associations. c , d Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. e Results given by Scissor under different \(\alpha\) values. f F1 scores and g FSC for SCIPAC and Scissor under different parameter values. h Cell differentiation paths. The four paths have the same starting location, which is in the center, but different ending locations. This can be considered as a progenitor cell type differentiating into four specialized cell types. i Cell differentiation steps. These steps are used to create four stages, each containing 500 steps. Thus, this plot of differentiation steps can also be viewed as the plot of true association strengths. j , k Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution
The association strengths and p -values given by SCIPAC under the default resolution are illustrated in Fig. 2 c, d, respectively. Results under several other resolutions are given in Additional file 1: Fig. S4 and S5. Again, we find that SCIPAC gives highly consistent results under different resolutions. SCIPAC successfully identifies three out of the four truly associated cell types. For the other truly associated cell type, cell type 1, SCIPAC correctly recognizes its association with the phenotype as negative, although the p -values are not significant enough. The F1 score is 0.85, and the FSC is greater than 0.99, as shown in Fig. 2 f, g.
The results of Scissor under four different \(\alpha\) values are given in Fig. 2 e. (More shown in Additional file 1: Fig. S6.) Under this highly challenging simulation scheme, Scissor can only identify one out of four truly associated cell types. Its F1 score is below 0.4.
Simulation scheme III
This simulation scheme is to assess the performance of SCIPAC for ordinal phenotypes. We simulate cells along four cell-differentiation paths with the same starting location but different ending locations, as shown in Fig. 2 h. These cells can be considered as a progenitor cell population differentiating into four specialized cell types. In Fig. 2 i, the “step” reflects their position in the differentiation path, with step 0 meaning the start and step 2000 meaning the end of the differentiation. Then, the “stage” is generated according to the step: cells in steps 0 \(\sim\) 500, 501 \(\sim\) 1000, 1001 \(\sim\) 1500, and 1501 \(\sim\) 2000 are assigned to stages I, II, III, and IV, respectively. This stage is treated as the ordinal phenotype. Under this simulation scheme, Fig. 2 i also gives the actual associations, and all cells are associated with the phenotype.
The results of SCIPAC under the default resolution are shown in Fig. 2 j, k. Clearly, the associations SCIPAC identifies are highly consistent with the truth. Particularly, it successfully identifies the cells in the center as early-stage cells and most cells at the end of branches as last-stage cells. The results of SCIPAC under other resolutions are given in Additional file 1: Fig. S7 and S8, which are highly consistent. Scissor does not work with ordinal phenotypes; thus, no results are reported here.
Performance in real data
We consider four real datasets: a prostate cancer dataset, a breast cancer dataset, a lung cancer dataset, and a muscular dystrophy dataset. The bulk RNA-seq data of the three cancer datasets are obtained from the TCGA database, and that of the muscular dystrophy dataset is obtained from a published paper [ 28 ]. A detailed description of these datasets is given in Additional file 1. We will use these datasets to assess the performance of SCIPAC on different types of phenotypes. The cell type information (i.e., which cell belongs to which cell type) is available for the first three datasets, but we ignore this information so that we can make a fair comparison with Scissor, which cannot utilize this information.
Prostate cancer data with a binary phenotype
We use the single-cell expression of 8,700 cells from prostate-cancer tumors sequenced by [ 29 ]. The cell types of these cells are known and given in Fig. 3 a. The bulk data comprises 550 TCGA-PRAD (prostate adenocarcinoma) samples with phenotype (cancer vs. normal) information. Here the phenotype is cancer, and it is binary: present or absent.
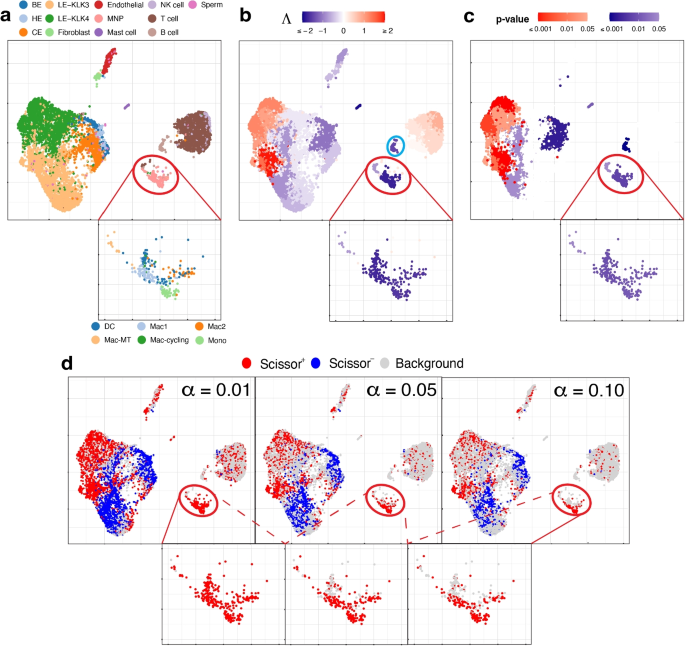
UMAP visualization of the prostate cancer data, with a zoom-in view for the red-circled region (cell type MNP). a True cell types. BE, HE, and CE stand for basal, hillock, club epithelial cells, LE-KLK3 and LE-KLK4 stand for luminal epithelial cells with high levels of kallikrein related peptidase 3 and 4, and MNP stands for mononuclear phagocytes. In the zoom-in view, the sub-types of MNP cells are given. b Association strengths \(\Lambda\) given by SCIPAC under the default resolution. The cyan-circled cells are B cells, which are estimated by SCIPAC as negatively associated with cancer but estimated by Scissor as Scissor+ or null. c p -values given by SCIPAC. The MNP cell type, which is red-circled in the plot, is estimated by SCIPAC to be strongly negatively associated with cancer but estimated by Scissor to be positively associated with cancer. d Results given by Scissor under different \(\alpha\) values
Results from SCIPAC with the default resolution are shown in Fig. 3 b, c (results with other resolutions, given in Additional file 1: Fig. S9 and S10, are highly consistent with results here.) Compared with results from Scissor, shown in Fig. 3 d, results from SCIPAC again show three advantages. First, results from SCIPAC are richer and more comprehensive. SCIPAC gives estimated associations and the corresponding p -values, and the estimated associations are quantitative (shown in Fig. 3 b as different shades to the red or blue color) instead of discrete (shown in Fig. 3 d as a uniform shade to the red, blue, or light gray color). Second, SCIPAC’s results can be easier to interpret as the red and blue colors are more block-wise instead of scattered. Third, unlike Scissor, which produces multiple sets of results varying based on the parameter \(\alpha\) —a parameter without a default value or tuning guidance—typically, a single set of results from SCIPAC under its default settings suffices.
Comparing the results from our SCIPAC method with those from Scissor is non-trivial, as the latter’s outcomes are scattered and include multiple sets. We propose the following solutions to summarize the inferred association of a known cell type with the phenotype using a specific method (Scissor under a specific \(\alpha\) value, or SCIPAC with the default setting). We first calculate the proportion of cells in this cell type identified as Scissor+ (by Scissor at a specific \(\alpha\) value) or as significantly positively associated (by SCIPAC), denoted by \(p_{+}\) . We also calculate the proportion of all cells, encompassing any cell type, which are identified as Scissor+ or significantly positively associated, serving as the average background strength, denoted by \(p_{a}\) . Then, we compute the log odds ratio for this cell type to be positively associated with the phenotype compared to the background, represented as:
Similarly, the log odds ratio for the cell type to be negatively associated with the phenotype, \(\rho _-\) , is computed in a parallel manner.
For SCIPAC, a cell type is summarized as positively associated with the phenotype if \(\rho _+ \ge 1\) and \(\rho _- < 1\) and negatively associated if \(\rho _- \ge 1\) and \(\rho _+ < 1\) . If neither condition is met, the association is inconclusive. For Scissor, we apply it under six different \(\alpha\) values: 0.01, 0.05, 0.10, 0.15, 0.20, and 0.25. A cell type is summarized as positively associated with the phenotype if \(\rho _+ \ge 1\) and \(\rho _- < 1\) in at least four of these \(\alpha\) values and negatively associated if \(\rho _- \ge 1\) and \(\rho _+ < 1\) in at least four \(\alpha\) values. If these criteria are not met, the association is deemed inconclusive. The above computation of log odds ratios and the determination of associations are performed only on cell types that each compose at least 1% of the cell population, to ensure adequate power.
For the prostate cancer data, the log odds ratios for each cell type using each method are presented in Tables S1 and S2. The final associations determined for each cell type are summarized in Table S3. In the last column of this table, we also indicate whether the conclusions drawn from SCIPAC and Scissor are consistent or not.
We find that SCIPAC’s results agree with Scissor on most cell types. However, there are three exceptions: mononuclear phagocytes (MNPs), B cells, and LE-KLK4.
MNPs are red-circled and zoomed in in each sub-figure of Fig. 3 . Most cells in this cell type are colored red in Fig. 3 d but colored dark blue in Fig. 3 b. In other words, while Scissor determines that this cell type is Scissor+, SCIPAC makes the opposite inference. Moreover, SCIPAC is confident about its judgment by giving small p -values, as shown in Fig. 3 c. To see which inference is closer to the biological fact is not easy, as biologically MNPs contain a number of sub-types that each have different functions [ 30 , 31 ]. Fortunately, this cell population has been studied in detail in the original paper that generated this dataset [ 29 ], and the sub-type information of each cell is provided there: this MNP population contains six sub-types, which are dendritic cells (DC), M1 macrophages (Mac1), metallothionein-expressing macrophages (Mac-MT), M2 macrophages (Mac2), proliferating macrophages (Mac-cycling), and monocytes (Mono), as shown in the zoom-in view of Fig. 3 a. Among these six sub-types, DC, Mac1, and Mac-MT are believed to inhibit cancer development and can serve as targets in cancer immunotherapy [ 29 ]; they compose more than 60% of all MNP cells in this dataset. SCIPAC makes the correct inference on this majority of MNP cells. Another cell type, Mac2, is reported to promote tumor development [ 32 ], but it only composes less than \(15\%\) of the MNPs. How the other two cell types, Mac-cycling and Mono, are associated with cancer is less studied. Overall, the results given by SCIPAC are more consistent with the current biological knowledge.
B cells are cyan-circled in Fig. 3 b. B cells are generally believed to have anti-tumor activity by producing tumor-reactive antibodies and forming tertiary lymphoid structures [ 29 , 33 ]. This means that B cells are likely to be negatively associated with cancer. SCIPAC successfully identifies this negative association, while Scissor fails.
LE-KLK4, a subtype of cancer cells, is thought to be positively associated with the tumor phenotype [ 29 ]. SCIPAC successfully identified this positive association, in contrast to Scissor, which failed to do so (in the figure, a proportion of LE-KLK4 cells are identified as Scissor+, especially under the smallest \(\alpha\) value; however, this proportion is not significantly higher than the background Scissor+ level under the majority of \(\alpha\) values).
In summary, across all three cell types, the results from SCIPAC appear to be more consistent with current biological knowledge. For more discussions regarding this dataset, refer to Additional file 1.
Breast cancer data with an ordinal phenotype
The scRNA-seq data for breast cancer are from [ 34 ], and we use the 19,311 cells from the five HER2+ tumor tissues. The true cell types are shown in Fig. 4 a. The bulk data include 1215 TCGA-BRCA samples with information on the cancer stage (I, II, III, or IV), which is treated as an ordinal phenotype.
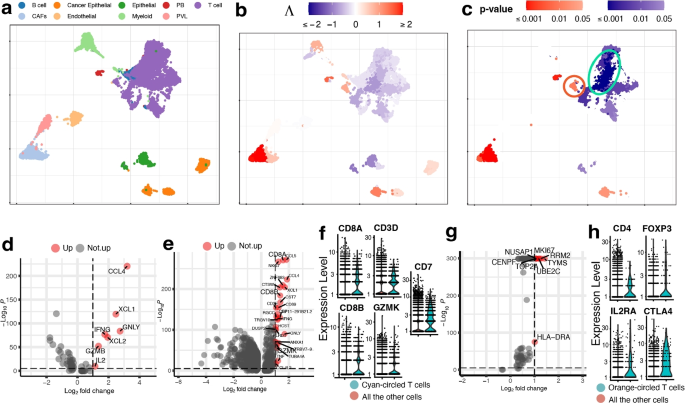
UMAP visualization of the breast cancer data. a True cell types. CAFs stand for cancer-associated fibroblasts, PB stands for plasmablasts and PVL stands for perivascular-like cells. b , c Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. Cyan-circled are a group of T cells that are estimated by SCIPAC to be most significantly associated with the cancer stage in the negative direction, and orange-circled are a group of T cells that are estimated by SCIPAC to be significantly positively associated with the cancer stage. d DE analysis of the cyan-circled T cells vs. all the other T cells. e DE analysis of the cyan-circled T cells vs. all the other cells. f Expression of CD8+ T cell marker genes in the cyan-circled cells and all the other cells. g DE analysis of the orange-circled T cells vs. all the other cells. h Expression of regulatory T cell marker genes in the orange-circled cells and all the other cells
Association strengths and p -values given by SCIPAC under the default resolution are shown in Fig. 4 b, c. Results under other resolutions are given in Additional file 1: Fig. S11 and S12, and again they are highly consistent with results under the default resolution. We do not present the results from Scissor, as Scissor does not take ordinal phenotypes.
In the SCIPAC results, cells that are most strongly and statistically significantly associated with the phenotype in the positive direction are the cancer-associated fibroblasts (CAFs). This finding agrees with the literature: CAFs contribute to therapy resistance and metastasis of cancer cells via the production of secreted factors and direct interaction with cancer cells [ 35 ], and they are also active players in breast cancer initiation and progression [ 36 , 37 , 38 , 39 ]. Another large group of cells identified as positively associated with the phenotype is the cancer epithelial cells. They are malignant cells in breast cancer tissues and are thus expected to be associated with severe cancer stages.
Of the cells identified as negatively associated with severe cancer stages, a large portion of T cells is the most noticeable. Biologically, T cells contain many sub-types, including CD4+, CD8+, regulatory T cells, and more, and their functions are diverse in the tumor microenvironment [ 40 ]. To explore SCIPAC’s discoveries, we compare T cells that are identified as most statistically significant, with p -values \(< 10^{-6}\) and circled in Fig. 4 d, with the other T cells. Differential expression (DE) analysis (details about DE analysis and other analyses are given in Additional file 1) identifies seven genes upregulated in these most significant T cells. Of these seven genes, at least five are supported by the literature: CCL4, XCL1, IFNG, and GZMB are associated with CD8+ T cell infiltration; they have been shown to have anti-tumor functions and are involved in cancer immunotherapy [ 41 , 42 , 43 ]. Also, IL2 has been shown to serve an important role in combination therapies for autoimmunity and cancer [ 44 ]. We also perform an enrichment analysis [ 45 ], in which a pathway called Myc stands out with a \(\textit{p}\text{-value}<10^{-7}\) , much smaller than all other pathways. Myc is downregulated in the T cells that are identified as most negatively associated with cancer stage progress. This agrees with current biological knowledge about this pathway: Myc is known to contribute to malignant cell transformation and tumor metastasis [ 46 , 47 , 48 ].
On the above, we have compared T cells that are most significantly associated with cancer stages in the negative direction with the other T cells using DE and pathway analysis, and the results could suggest that these cells are tumor-infiltrated CD8+ T cells with tumor-inhibition functions. To check this hypothesis, we perform DE analysis of these cells against all other cells (i.e., the other T cells and all the other cell types). The DE genes are shown in Fig. 4 e. It can be noted that CD8+ T cell marker genes such as CD8A, CD8B, and GZMK are upregulated. We further obtain CD8+ T cell marker genes from CellMarker [ 49 ] and check their expression, as illustrated in Fig. 4 f. Marker genes CD8A, CD8B, CD3D, GZMK, and CD7 show significantly higher expression in these T cells. This again supports our hypothesis that these cells are tumor-infiltrated CD8+ T cells that have anti-tumor functions.
Interestingly, not all T cells are identified as negatively associated with severe cancer stages; a group of T cells is identified as positively associated, as circled in Fig. 4 c. To explore the function of this group of T cells, we perform DE analysis of these T cells against the other T cells. The DE genes are shown in Fig. 4 g. Based on the literature, six out of eight over-expressed genes are associated with cancer development. The high expression of NUSAP1 gene is associated with poor patient overall survival, and this gene also serves as a prognostic factor in breast cancer [ 50 , 51 , 52 ]. Gene MKI67 has been treated as a candidate prognostic prediction for cancer proliferation [ 53 , 54 ]. The over-expression of RRM2 has been linked to higher proliferation and invasiveness of malignant cells [ 55 , 56 ], and the upregulation of RRM2 in breast cancer suggests it to be a possible prognostic indicator [ 57 , 58 , 59 , 60 , 61 , 62 ]. The high expression of UBE2C gene always occurs in cancers with a high degree of malignancy, low differentiation, and high metastatic tendency [ 63 ]. For gene TOP2A, it has been proposed that the HER2 amplification in HER2 breast cancers may be a direct result of the frequent co-amplification of TOP2A [ 64 , 65 , 66 ], and there is a high correlation between the high expressions of TOP2A and the oncogene HER2 [ 67 , 68 ]. Gene CENPF is a cell cycle-associated gene, and it has been identified as a marker of cell proliferation in breast cancers [ 69 ]. The over-expression of these genes strongly supports the correctness of the association identified by SCIPAC. To further validate this positive association, we perform DE analysis of these cells against all the other cells. We find that the top marker genes obtained from CellMarker [ 49 ] for the regulatory T cells, which are known to be immunosuppressive and promote cancer progression [ 70 ], are over-expressed with statistical significance, as shown in Fig. 4 h. This finding again provides strong evidence that the positive association identified by SCIPAC for this group of T cells is correct.
Lung cancer data with survival information
The scRNA-seq data for lung cancer are from [ 71 ], and we use two lung adenocarcinoma (LUAD) patients’ data with 29,888 cells. The true cell types are shown in Fig. 5 a. The bulk data consist of 576 TCGA-LUAD samples with survival status and time.
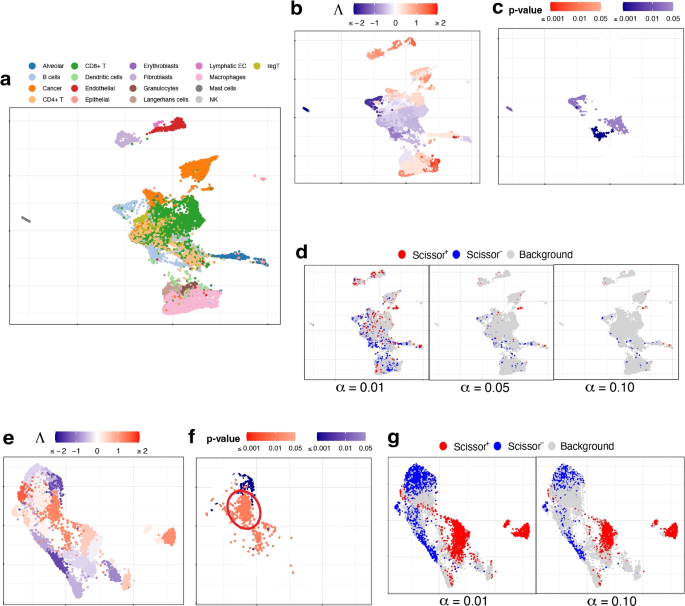
UMAP visualization of a–d the lung cancer data and e–g the muscular dystrophy data. a True cell types. b , c Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. d Results given by Scissor under different \(\alpha\) values. e , f Association strengths \(\Lambda\) and p -values given by SCIPAC under the default resolution. Circled are a group of cells that are identified by SCIPAC as significantly positively associated with the disease but identified by Scissor as null. g Results given by Scissor under different \(\alpha\) values
Association strengths and p -values given by SCIPAC are given in Fig. 5 b, c (results under other resolutions are given in Additional file 1: Fig. S13 and S14). In Fig. 5 c, most cells with statistically significant associations are CD4+ T cells or B cells. These associations are negative, meaning that the abundance of these cells is associated with a reduced death rate, i.e., longer survival time. This agrees with the literature: CD4+ T cells primarily mediate anti-tumor immunity and are associated with favorable prognosis in lung cancer patients [ 72 , 73 , 74 ]; B cells also show anti-tumor functions in all stages of human lung cancer development and play an essential role in anti-tumor responses [ 75 , 76 ].
The results by Scissor under different \(\alpha\) values are shown in Fig. 5 d. The highly scattered Scissor+ and Scissor− cells make identifying and interpreting meaningful phenotype-associated cell groups difficult.
Muscular dystrophy data with a binary phenotype
This dataset contains cells from four facioscapulohumeral muscular dystrophy (FSHD) samples and two control samples [ 77 ]. We pool all the 7047 cells from these six samples together. The true cell types of these cells are unknown. The bulk data consists of 27 FSHD patients and eight controls from [ 28 ]. Here the phenotype is FSHD, and it is binary: present or absent.
The results of SCIPAC with the default resolution are given in Fig. 5 e, f. Results under other resolutions are highly similar (shown in Additional file 1: Fig. S15 and S16). For comparison, results given by Scissor under different \(\alpha\) values are presented in Fig. 5 g. The agreements between the results of SCIPAC and Scissor are clear. For example, both methods identify cells located at the top and lower left part of UMAP plots to be negatively associated with FSHD, and cells located at the center and right parts of UMAP plots to be positively associated. However, the discrepancies in their results are also evident. The most pronounced one is a large group of cells (circled in Fig. 5 f) that are identified by SCIPAC as significantly positively associated but are completely ignored by Scissor. Checking into this group of cells, we find that over 90% (424 out of 469) come from the FSHD patients, and less than 10% come from the control samples. However, cells from FSHD patients only compose 73% (5133) of all the 7047 cells. This statistically significant ( p -value \(<10^{-15}\) , Fisher’s exact test) over-representation (odds ratio = 3.51) suggests that the positive association identified SCIPAC is likely to be correct.
SCIPAC is computationally highly efficient. On an 8-core machine with 2.50 GHz CPU and 16 GB RAM, SCIPAC takes 7, 24, and 2 s to finish all the computation and give the estimated association strengths and p -values on the prostate cancer, lung cancer, and muscular dystrophy datasets, respectively. As a reference, Scissor takes 314, 539, and 171 seconds, respectively.
SCIPAC works with various phenotype types, including binary, continuous, survival, and ordinal. It can easily accommodate other types by using a proper regression model with a systematic component in the form of Eq. 3 (see the “ Methods ” section). For example, a Poisson or negative binomial log-linear model can be used if the phenotype is a count (i.e., non-negative integer).
In SCIPAC’s definition of association, a cell type is associated with the phenotype if increasing the proportion of this cell type leads to a change of probability of the phenotype occurring. The strength of association represents the extent of the increase or decrease in this probability. In the case of binary-response, this change is measured by the log odds ratio. For example, if the association strength of cell type A is twice that of cell type B, increasing cell type A by a certain proportion leads to twice the amount of change in the log odds ratio of having the phenotype compared to increasing cell type B by the same proportion. The association strength under other types of phenotypes can be interpreted similarly, with the major difference lying in the measure of change in probability. For quantitative, ordinal, and survival outcomes, the difference in the quantitative outcome, log odds ratio of the right-tail probability, and log hazard ratio respectively are used. Despite the differences in the exact form of the association strength under different types of phenotypes, the underlying concept remains the same: a larger (absolute value of) association strength indicates that the same increase/decrease in a cell type leads to a larger change in the occurrence of the phenotype.
As SCIPAC utilizes both bulk RNA-seq data with phenotype and single-cell RNA-seq data, the estimated associations for the cells are influenced by the choice of the bulk data. Although different bulk data can yield varying estimations of the association for the same single cells, the estimated associations appear to be reasonably robust even when minor changes are made to the bulk data. See Additional file 1 for further discussions.
When using the Louvain algorithm in the Seurat package to cluster cells, SCIPAC’s default resolution is 2.0, larger than the default setting of Seurat. This allows for the identification of potential subtypes within the major cell type and enables the estimation of individual association strengths. Consequently, a more detailed and comprehensive description of the association between single cells and the phenotype can be obtained by SCIPAC.
When applying SCIPAC to real datasets, we made a deliberate choice to disregard the cell annotation provided by the original publications and instead relied on the inferred cell clusters produced by the Louvain algorithm. We made this decision for several reasons. Firstly, we aimed to ensure a fair comparison with Scissor, as it does not utilize cell-type annotations. Secondly, the original annotation might not be sufficiently comprehensive or detailed. Presumed cell types could potentially encompass multiple subtypes, each of which may exhibit distinct associations with the phenotype under investigation. In such cases, employing the Louvain algorithm with a relatively high resolution, which is the default setting in SCIPAC, enables us to differentiate between these subtypes and allows SCIPAC to assign varying association strengths to each subtype.
SCIPAC fits the regression model using the elastic net, a machine-learning algorithm that maximizes a penalized version of the likelihood. The elastic net can be replaced by other penalized estimates of regression models, such as SCAD [ 78 ], without altering the rest of the SCIPAC algorithm. The combination of a regression model and a penalized estimation algorithm such as the elastic net has shown comparable or higher prediction power than other sophisticated methods such as random forests, boosting, or neural networks in numerous applications, especially for gene expression data [ 79 ]. However, there can still be datasets where other models have higher prediction power. It will be future work to incorporate these models into SCIPAC.
The use of metacells is becoming an efficient way to handle large single-cell datasets [ 80 , 81 , 82 , 83 ]. Conceptually, SCIPAC can incorporate metacells and their representatives as an alternative to its default setting of using cell clusters/types and their centroids. We have explored this aspect using metacells provided by SEACells [ 81 ]. Details are given in Additional file 1. Our comparative analysis reveals that combining SCIPAC with SEACells results in significantly reduced performance compared to using SCIPAC directly on original single-cell data. The primary reason for this appears to be the subpar performance of SEACells in cell grouping, especially when contrasted with the Louvain algorithm. Given these findings, we do not suggest using metacells provided by SEACells for SCIPAC applications in the current stage.
Conclusions
SCIPAC is a novel algorithm for studying the associations between cells and phenotypes. Compared to the previous algorithm, SCIPAC gives a much more detailed and comprehensive description of the associations by enabling a quantitative estimation of the association strength and by providing a quality control—the p -value. Underlying SCIPAC are a general statistical model that accommodates virtually all types of phenotypes, including ordinal (and potentially count) phenotypes that have never been considered before, and a concise and closed-form mathematical formula that quantifies the association, which minimizes the computational load. The mathematical conciseness also largely frees SCIPAC from parameter tuning. The only parameter (i.e., the resolution) barely changes the results given by SCIPAC. Overall, compared with its predecessor, SCIPAC represents a substantially more capable software by being much more informative, versatile, robust, and user-friendly.
The improvement in accuracy is also remarkable. In simulated data, SCIPAC achieves high power and low false positives, which is evident from the UMAP plot, F1 score, and FSC score. In real data, SCIPAC gives results that are consistent with current biological knowledge for cell types whose functions are well understood. For cell types whose functions are less studied or more multifaceted, SCIPAC gives support to certain biological hypotheses or helps identify/discover cell sub-types.
SCIPAC’s identification of cell-phenotype associations closely follows its definition of association: when increasing the fraction of a cell type increases (or decreases) the probability for a phenotype to be present, this cell type is positively (or negatively) associated with the phenotype.
The increase of the fraction of a cell type
For a bulk sample, let vector \(\varvec{G} \in \mathbb {R}^p\) be its expression profile, that is, its expression on the p genes. Suppose there are K cell types in the tissue, and let \(\varvec{g}_{k}\) be the representative expression of the k ’th cell type. Usually, people assume that \(\varvec{G}\) can be decomposed by
where \(\gamma _{k}\) is the proportion of cell type k in the bulk tissue, with \(\sum _{k = 1}^{K}\gamma _{k} = 1\) . This equation links the bulk and single-cell expression data.
Now consider increasing cells from cell type k by \(\Delta \gamma\) proportion of the original number of cells. Then, the new proportion of cell type k becomes \(\frac{\gamma _{k} + \Delta \gamma }{1 + \Delta \gamma }\) , and the new proportion of cell type \(j \ne k\) becomes \(\frac{\gamma _{j}}{1 + \Delta \gamma }\) (note that the new proportions of all cell types should still add up to 1). Thus, the bulk expression profile with the increase of cell type k becomes
Plugging Eq. 1 , we get
Interestingly, this expression of \(\varvec{G}^*\) does not include \(\gamma _{1}, \ldots , \gamma _{K}\) . This means that there is no need actually to compute \(\gamma _{1}, \ldots , \gamma _{K}\) in Eq. 1 , which could otherwise be done using a cell-type-decomposition software, but an accurate and robust decomposition is non-trivial [ 84 , 85 , 86 ]. See Additional file 1 for a more in-depth discussion on the connections of SCIPAC with decomposition/deconvolution.
The change in chance of a phenotype
In this section, we consider how the increase in the fraction of a cell type will change the chance for a binary phenotype such as cancer to occur. Other types of phenotypes will be considered in the next section.
Let \(\pi (\varvec{G})\) be the chance of an individual with gene expression profile \(\varvec{G}\) for this phenotype to occur. We assume a logistic regression model to describe the relationship between \(\pi (\varvec{G})\) and \(\varvec{G}\) :
here the left-hand side is the log odds of \(\pi (\varvec{G})\) , \(\beta _{0}\) is the intercept, and \(\varvec{\beta }\) is a length- p vector of coefficients. In the section after the next, we will describe how we obtain \(\beta _{0}\) and \(\varvec{\beta }\) from the data.
When increasing cells from cell type k by \(\Delta \gamma\) , \(\varvec{G}\) becomes \(\varvec{G}^*\) in Eq. 3 . Plugging Eq. 2 , we get
We further take the difference between Eqs. 4 and 3 and get
The left-hand side of this equation is the log odds ratio (i.e., the change of log odds). On the right-hand side, \(\frac{\Delta \gamma }{1 + \Delta \gamma }\) is an increasing function with respect to \(\Delta \gamma\) , and \(\varvec{\beta }^T(\varvec{g}_{k} - \varvec{G})\) is independent of \(\Delta \gamma\) . This indicates that given any specific \(\Delta \gamma\) , the log odds ratio under over-representation of cell type k is proportional to
\(\lambda _k\) describes the strength of the effect of increasing cell type k to a bulk sample with expression profile \(\varvec{G}\) . Given the presence of numerous bulk samples, employing multiple \(\lambda _k\) ’s could be cumbersome and obscure the overall effect of a particular cell type. To concisely summarize the association of cell type k , we propose averaging their effects. The average effect on all bulk samples can be obtained by
where \(\bar{\varvec{G}}\) is the average expression profile of all bulk samples.
\(\Lambda _k\) gives an overall impression of how strong the effect is when cell type k over-represents to the probability for the phenotype to be present. Its sign represents the direction of the change: a positive value means an increase in probability, and a negative value means a decrease in probability. Its absolute value represents the strength of the effect. In SCIPAC, we call \(\Lambda _k\) the association strength of cell type k and the phenotype.
Note that this derivation does not involve likelihood, although the computation of \(\varvec{\beta }\) does. Here, it serves more as a definitional approach.
Definition of the association strength for other types of phenotype
Our definition of \(\Lambda _k\) relies on vector \(\varvec{\beta }\) . In the case of a binary phenotype, \(\varvec{\beta }\) are the coefficients of a logistic regression that describes a linear relationship between the expression profile and the log odds of having the phenotype, as shown in Eq. 3 . For other types of phenotype, \(\varvec{\beta }\) can be defined/computed similarly.
For a quantitative (i.e., continuous) phenotype, an ordinary linear regression can be used, and the left-hand side of Eq. 3 is changed to the quantitative value of the phenotype.
For a survival phenotype, a Cox proportional hazards model can be used, and the left-hand side of Eq. 3 is changed to the log hazard ratio.
For an ordinal phenotype, we use a proportional odds model
where \(j \in \{1, 2, ..., (J - 1)\}\) and J is the number of ordinal levels. It should be noted that here we use the right-tail probability \(\Pr (Y_{i} \ge j + 1 | X)\) instead of the commonly used cumulative probability (left-tail probability) \(\Pr (Y_{i} \le j | X)\) . Such a change makes the interpretation consistent with other types of phenotypes: in our model, a larger value on the right-hand side indicates a larger chance for \(Y_{i}\) to have a higher level, which in turn guarantees that the sign of the association strength defined according to this \(\varvec{\beta }\) has the usual meaning: a positive \(\Lambda _k\) value means a positive association with the phenotype-using the cancer stage as an example. A positive \(\Lambda _k\) means the over-representation of cell type k increases the chance of a higher cancer stage. In contrast, using the commonly used cumulative probability leads to a counter-intuitive, reversed interpretation.
Computation of the association strength in practice
In practice, \(\varvec{\beta }\) in Eq. 3 needs to be learned from the bulk data. By default, SCIPAC uses the elastic net, a popular and powerful penalized regression method:
In this model, \(l(\beta _{0}, \varvec{\beta })\) is a log-likelihood of the linear model (i.e., logistic regression for a binary phenotype, ordinary linear regression for a quantitative phenotype, Cox proportional odds model for a survival phenotype, and proportional odds model for an ordinal phenotype). \(\alpha\) is a number between 0 and 1, denoting a combination of \(\ell _1\) and \(\ell _2\) penalties, and \(\lambda\) is the penalty strength. SCIPAC fixes \(\alpha\) to be 0.4 (see Additional file 1 for discussions on this choice) and uses 10-fold cross-validation to decide \(\lambda\) automatically. This way, they do not become hyperparameters.
In SCIPAC, the fitting and cross-validation of the elastic net are done by calling the ordinalNet [ 87 ] R package for the ordinal phenotype and by calling the glmnet R package [ 88 , 89 , 90 , 91 ] for other types of phenotypes.
The computation of the association strength, as defined by Eq. 7 , does not only require \(\varvec{\beta }\) , but also \(\varvec{g}_k\) and \(\bar{\varvec{G}}\) . \(\bar{\varvec{G}}\) is simply the average expression profile of all bulk samples. On the other hand, \(\varvec{g}_k\) requires knowing the cell type of each cell. By default, SCIPAC does not assume this information to be given, and it uses the Louvain clustering implemented in the Seurat [ 24 , 25 ] R package to infer it. This clustering algorithm has one tuning parameter called “resolution.” SCIPAC sets its default value as 2.0, and the user can use other values. With the inferred or given cell types, \(\varvec{g}_k\) is computed as the centroid (i.e., the mean expression profile) of cells in cluster k .
Given \(\varvec{\beta }\) , \(\bar{\varvec{G}}\) , and \(\varvec{g}_k\) , the association strength can be computed using Eq. 7 . Knowing the association strength for each cell type and the cell-type label for each cell, we also know the association strength for every single cell. In practice, we standardize the association strengths for all cells. That is, we compute the mean and standard deviation of the association strengths of all cells and use them to centralize and scale the association strength, respectively. We have found such standardization makes SCIPAC more robust to the possible unbalance in sample size of bulk data in different phenotype groups.
Computation of the p -value
SCIPAC uses non-parametric bootstrap [ 92 ] to compute the standard deviation and hence the p -value of the association. Fifty bootstrap samples, which are believed to be enough to compute the standard error of most statistics [ 93 ], are generated for the bulk expression data, and each is used to compute (standardized) \(\Lambda\) values for all the cells. For cell i , let its original \(\Lambda\) values be \(\Lambda _i\) , and the bootstrapped values be \(\Lambda _i^{(1)}, \ldots , \Lambda _i^{(50)}\) . A z -score is then computed using
and then the p -value is computed according to the cumulative distribution function of the standard Gaussian distribution. See Additional file 1 for more discussions on the calculation of p -value.
Availability of data and materials
The simulated datasets [ 94 ] under three schemes are available at Zenodo with DOI 10.5281/zenodo.11013320 [ 95 ]. The SCIPAC package is available at GitHub website https://github.com/RavenGan/SCIPAC under the MIT license [ 96 ]. The source code of SCIPAC is also deposited at Zenodo with DOI 10.5281/zenodo.11013696 [ 97 ]. A vignette of the R package is available on the GitHub page and in the Additional file 2. The prostate cancer scRNA-seq data is obtained from the Prostate Cell Atlas https://www.prostatecellatlas.org [ 29 ]; the scRNA-seq data for the breast cancer are from the Gene Expression Omnibus (GEO) under accession number GSE176078 [ 34 , 98 ]; the scRNA-seq data for the lung cancer are from E-MTAB-6149 [ 99 ] and E-MTAB-6653 [ 71 , 100 ]; the scRNA-seq data for facioscapulohumeral muscular dystrophy data are from the GEO under accession number GSE122873 [ 101 ]. The bulk RNA-seq data are obtained from the TCGA database via TCGAbiolinks (ver. 2.25.2) R package [ 102 ]. More details about the simulated and real scRNA-seq and bulk RNA-seq data can be found in the Additional file 1.
Yofe I, Dahan R, Amit I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat Med. 2020;26(2):171–7.
Article CAS PubMed Google Scholar
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–45.
Fan J, Slowikowski K, Zhang F. Single-cell transcriptomics in cancer: computational challenges and opportunities. Exp Mol Med. 2020;52(9):1452–65.
Article CAS PubMed PubMed Central Google Scholar
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14.
Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360(6385):176–82.
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8(1):1–12.
Article Google Scholar
Abdelaal T, Michielsen L, Cats D, Hoogduin D, Mei H, Reinders MJ, et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20(1):1–19.
Article CAS Google Scholar
Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15(6):e8746.
Article PubMed PubMed Central Google Scholar
Guo H, Li J. scSorter: assigning cells to known cell types according to marker genes. Genome Biol. 2021;22(1):1–18.
Pliner HA, Shendure J, Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16(10):983–6.
Zhang AW, O’Flanagan C, Chavez EA, Lim JL, Ceglia N, McPherson A, et al. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat Methods. 2019;16(10):1007–15.
Zhang Z, Luo D, Zhong X, Choi JH, Ma Y, Wang S, et al. SCINA: a semi-supervised subtyping algorithm of single cells and bulk samples. Genes. 2019;10(7):531.
Johnson TS, Wang T, Huang Z, Yu CY, Wu Y, Han Y, et al. LAmbDA: label ambiguous domain adaptation dataset integration reduces batch effects and improves subtype detection. Bioinformatics. 2019;35(22):4696–706.
Ma F, Pellegrini M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics. 2020;36(2):533–8.
Tan Y, Cahan P. SingleCellNet: a computational tool to classify single cell RNA-Seq data across platforms and across species. Cell Syst. 2019;9(2):207–13.
Salcher S, Sturm G, Horvath L, Untergasser G, Kuempers C, Fotakis G, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40(12):1503–20.
Good Z, Sarno J, Jager A, Samusik N, Aghaeepour N, Simonds EF, et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med. 2018;24(4):474–83.
Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177(5):1330–45.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–20.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disc. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1.
Sun D, Guan X, Moran AE, Wu LY, Qian DZ, Schedin P, et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol. 2022;40(4):527–38.
Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008(10):P10008.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902.
Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol. 2005;67(2):301–20.
McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2018. arXiv preprint arXiv:1802.03426 .
Wong CJ, Wang LH, Friedman SD, Shaw D, Campbell AE, Budech CB, et al. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum Mol Genet. 2020;29(6):1030–43.
Tuong ZK, Loudon KW, Berry B, Richoz N, Jones J, Tan X, et al. Resolving the immune landscape of human prostate at a single-cell level in health and cancer. Cell Rep. 2021;37(12):110132.
Hume DA. The mononuclear phagocyte system. Curr Opin Immunol. 2006;18(1):49–53.
Hume DA, Ross IL, Himes SR, Sasmono RT, Wells CA, Ravasi T. The mononuclear phagocyte system revisited. J Leukoc Biol. 2002;72(4):621–7.
Raggi F, Bosco MC. Targeting mononuclear phagocyte receptors in cancer immunotherapy: new perspectives of the triggering receptor expressed on myeloid cells (TREM-1). Cancers. 2020;12(5):1337.
Largeot A, Pagano G, Gonder S, Moussay E, Paggetti J. The B-side of cancer immunity: the underrated tune. Cells. 2019;8(5):449.
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–47.
Fernández-Nogueira P, Fuster G, Gutierrez-Uzquiza Á, Gascón P, Carbó N, Bragado P. Cancer-associated fibroblasts in breast cancer treatment response and metastasis. Cancers. 2021;13(13):3146.
Ao Z, Shah SH, Machlin LM, Parajuli R, Miller PC, Rawal S, et al. Identification of cancer-associated fibroblasts in circulating blood from patients with metastatic breast cancer. Identification of cCAFs from metastatic cancer patients. Cancer Res. 2015;75(22):4681–7.
Arcucci A, Ruocco MR, Granato G, Sacco AM, Montagnani S. Cancer: an oxidative crosstalk between solid tumor cells and cancer associated fibroblasts. BioMed Res Int. 2016;2016. https://pubmed.ncbi.nlm.nih.gov/27595103/ .
Buchsbaum RJ, Oh SY. Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers. 2016;8(2):19.
Ruocco MR, Avagliano A, Granato G, Imparato V, Masone S, Masullo M, et al. Involvement of breast cancer-associated fibroblasts in tumor development, therapy resistance and evaluation of potential therapeutic strategies. Curr Med Chem. 2018;25(29):3414–34.
Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–93.
Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27(5):820–32.
Romero JM, Grünwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M, et al. A four-chemokine signature is associated with a T-cell-inflamed phenotype in primary and metastatic pancreatic cancer. Chemokines in Pancreatic Cancer. Clin Cancer Res. 2020;26(8):1997–2010.
Tamura R, Yoshihara K, Nakaoka H, Yachida N, Yamaguchi M, Suda K, et al. XCL1 expression correlates with CD8-positive T cells infiltration and PD-L1 expression in squamous cell carcinoma arising from mature cystic teratoma of the ovary. Oncogene. 2020;39(17):3541–54.
Hernandez R, Põder J, LaPorte KM, Malek TR. Engineering IL-2 for immunotherapy of autoimmunity and cancer. Nat Rev Immunol. 2022:22:1–15. https://pubmed.ncbi.nlm.nih.gov/35217787/ .
Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. BioRxiv. 2016:060012. https://www.biorxiv.org/content/10.1101/060012v3.abstract .
Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35.
Gnanaprakasam JR, Wang R. MYC in regulating immunity: metabolism and beyond. Genes. 2017;8(3):88.
Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, et al. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21(8):2921.
Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, et al. Cell Marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–8.
Chen L, Yang L, Qiao F, Hu X, Li S, Yao L, et al. High levels of nucleolar spindle-associated protein and reduced levels of BRCA1 expression predict poor prognosis in triple-negative breast cancer. PLoS ONE. 2015;10(10):e0140572.
Li M, Yang B. Prognostic value of NUSAP1 and its correlation with immune infiltrates in human breast cancer. Crit Rev TM Eukaryot Gene Expr. 2022;32(3). https://pubmed.ncbi.nlm.nih.gov/35695609/ .
Zhang X, Pan Y, Fu H, Zhang J. Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell proliferation and enhances susceptibility to epirubicin in invasive breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5 expression. Med Sci Monit: Int Med J Exp Clin Res. 2018;24:8553.
Geyer FC, Rodrigues DN, Weigelt B, Reis-Filho JS. Molecular classification of estrogen receptor-positive/luminal breast cancers. Adv Anat Pathol. 2012;19(1):39–53.
Karamitopoulou E, Perentes E, Tolnay M, Probst A. Prognostic significance of MIB-1, p53, and bcl-2 immunoreactivity in meningiomas. Hum Pathol. 1998;29(2):140–5.
Duxbury MS, Whang EE. RRM2 induces NF- \(\kappa\) B-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun. 2007;354(1):190–6.
Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, et al. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16(1):43–9.
Zhang H, Liu X, Warden CD, Huang Y, Loera S, Xue L, et al. Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer. 2014;14(1):1–16.
Putluri N, Maity S, Kommagani R, Creighton CJ, Putluri V, Chen F, et al. Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia. 2014;16(5):390–402.
Koleck TA, Conley YP. Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer Targets Ther. 2016;8:29.
Li Jp, Zhang Xm, Zhang Z, Zheng Lh, Jindal S, Liu Yj. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine. 2019;98(18). https://pubmed.ncbi.nlm.nih.gov/31045815/ .
Gong MT, Ye SD, Lv WW, He K, Li WX. Comprehensive integrated analysis of gene expression datasets identifies key anti-cancer targets in different stages of breast cancer. Exp Ther Med. 2018;16(2):802–10.
PubMed PubMed Central Google Scholar
Chen Wx, Yang Lg, Xu Ly, Cheng L, Qian Q, Sun L, et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep. 2019;39(4). https://pubmed.ncbi.nlm.nih.gov/30898978/ .
Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012;33(3):723–30.
Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, et al. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106(2):181–9.
Knoop AS, Knudsen H, Balslev E, Rasmussen BB, Overgaard J, Nielsen KV, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23(30):7483–90.
Tanner M, Isola J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, Malmstrom P, et al. Topoisomerase II \(\alpha\) gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24(16):2428–36.
Arriola E, Moreno A, Varela M, Serra JM, Falo C, Benito E, et al. Predictive value of HER-2 and topoisomerase II \(\alpha\) in response to primary doxorubicin in breast cancer. Eur J Cancer. 2006;42(17):2954–60.
Järvinen TA, Tanner M, Bärlund M, Borg Å, Isola J. Characterization of topoisomerase II \(\alpha\) gene amplification and deletion in breast cancer. Gene Chromosome Cancer. 1999;26(2):142–50.
Landberg G, Erlanson M, Roos G, Tan EM, Casiano CA. Nuclear autoantigen p330d/CENP-F: a marker for cell proliferation in human malignancies. Cytom J Int Soc Anal Cytol. 1996;25(1):90–8.
CAS Google Scholar
Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–8.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–89.
Bremnes RM, Busund LT, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, et al. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 2016;11(6):789–800.
Article PubMed Google Scholar
Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, et al. Objective measurement and clinical significance of TILs in non–small cell lung cancer. J Natl Cancer Inst. 2015;107(3):dju435.
Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy—new insights into old paradigms. Cancer Gene Ther. 2021;28(1):5–17.
Dieu-Nosjean MC, Goc J, Giraldo NA, Sautès-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 2014;35(11):571–80.
Wang Ss, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol. 2019;16(1):6–18.
van den Heuvel A, Mahfouz A, Kloet SL, Balog J, van Engelen BG, Tawil R, et al. Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Hum Mol Genet. 2019;28(7):1064–75.
Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc. 2001;96(456):1348–60.
Hastie T, Tibshirani R, Friedman JH, Friedman JH. The elements of statistical learning: data mining, inference, and prediction, vol. 2. New York: Springer; 2009.
Book Google Scholar
Baran Y, Bercovich A, Sebe-Pedros A, Lubling Y, Giladi A, Chomsky E, et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20(1):1–19.
Persad S, Choo ZN, Dien C, Sohail N, Masilionis I, Chaligné R, et al. SEACells infers transcriptional and epigenomic cellular states from single-cell genomics data. Nat Biotechnol. 2023;41:1–12. https://pubmed.ncbi.nlm.nih.gov/36973557/ .
Ben-Kiki O, Bercovich A, Lifshitz A, Tanay A. Metacell-2: a divide-and-conquer metacell algorithm for scalable scRNA-seq analysis. Genome Biol. 2022;23(1):100.
Bilous M, Tran L, Cianciaruso C, Gabriel A, Michel H, Carmona SJ, et al. Metacells untangle large and complex single-cell transcriptome networks. BMC Bioinformatics. 2022;23(1):336.
Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 2020;11(1):1–14.
Jin H, Liu Z. A benchmark for RNA-seq deconvolution analysis under dynamic testing environments. Genome Biol. 2021;22(1):1–23.
Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10(1):380.
Wurm MJ, Rathouz PJ, Hanlon BM. Regularized ordinal regression and the ordinalNet R package. 2017. arXiv preprint arXiv:1706.05003 .
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1.
Simon N, Friedman J, Hastie T. A blockwise descent algorithm for group-penalized multiresponse and multinomial regression. 2013. arXiv preprint arXiv:1311.6529 .
Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1.
Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, et al. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Ser B Stat Methodol. 2012;74(2):245–66.
Efron B. Bootstrap methods: another look at the jackknife. In: Breakthroughs in statistics. New York: Springer; 1992. pp. 569–593.
Efron B, Tibshirani RJ. An introduction to the bootstrap. London: CRC Press; 1994.
Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18(1):174.
Gan D, Zhu Y, Lu X, Li J. Simulated datasets used in SCIPAC analysis. Zenodo. 2024. https://doi.org/10.5281/zenodo.11013320 .
Gan D, Zhu Y, Lu X, Li J. SCIPAC R package. GitHub. 2024. https://github.com/RavenGan/SCIPAC . Accessed 24 Apr 2024.
Gan D, Zhu Y, Lu X, Li J. SCIPAC source code. Zenodo. 2024. https://doi.org/10.5281/zenodo.11013696 .
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Datasets. 2021. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176078 . Gene Expression Omnibus. Accessed 1 Oct 2022.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Datasets. 2018. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6149 . ArrayExpress. Accessed 24 July 2022.
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Datasets. 2018. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-6653 . ArrayExpress. Accessed 24 July 2022.
van den Heuvel A, Mahfouz A, Kloet SL, Balog J, van Engelen BG, Tawil R, et al. Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Datasets. 2019. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122873 . Gene Expression Omnibus. Accessed 13 Aug 2022.
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.
Download references
Review history
The review history is available as Additional file 3.
Peer review information
Veronique van den Berghe was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
This work is supported by the National Institutes of Health (R01CA280097 to X.L. and J.L, R01CA252878 to J.L.) and the DOD BCRP Breakthrough Award, Level 2 (W81XWH2110432 to J.L.).
Author information
Authors and affiliations.
Department of Applied and Computational Mathematics and Statistics, University of Notre Dame, Notre Dame, 46556, IN, USA
Dailin Gan & Jun Li
Department of Biological Sciences, Boler-Parseghian Center for Rare and Neglected Diseases, Harper Cancer Research Institute, Integrated Biomedical Sciences Graduate Program, University of Notre Dame, Notre Dame, 46556, IN, USA
Yini Zhu & Xin Lu
Tumor Microenvironment and Metastasis Program, Indiana University Melvin and Bren Simon Comprehensive Cancer Center, Indianapolis, 46202, IN, USA
You can also search for this author in PubMed Google Scholar
Contributions
J.L. conceived and supervised the study. J.L. and D.G. proposed the methods. D.G. implemented the methods and analyzed the data. D.G. and J.L. drafted the paper. D.G., Y.Z., X.L., and J.L. interpreted the results and revised the paper.
Corresponding author
Correspondence to Jun Li .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1. supplementary materials that include additional results and plots., additional file 2. a vignette of the scipac package., additional file 3. review history., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Gan, D., Zhu, Y., Lu, X. et al. SCIPAC: quantitative estimation of cell-phenotype associations. Genome Biol 25 , 119 (2024). https://doi.org/10.1186/s13059-024-03263-1
Download citation
Received : 30 January 2023
Accepted : 30 April 2024
Published : 13 May 2024
DOI : https://doi.org/10.1186/s13059-024-03263-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Phenotype association
- Single cell
- RNA sequencing
- Cancer research
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Methodology: Supplemental Document for the Green Sea Turtle (Chelonia mydas) Proposed Critical Habitat Designation in the Terrestrial Environment

As required by section 4(b)(2) of the Act, we use the best scientific data available to designate critical habitat. In accordance with the Act and our implementing regulations at 50 CFR 424.12(b), we review available information pertaining to the habitat requirements of the species and identify specific areas within the geographical area occupied by the species at the time of listing and any specific areas outside the geographical area occupied by the species to be considered for designation as critical habitat.

The green sea turtle grows to a maximum size of about 4 feet and a weight of 440 pounds. It has a heart-shaped shell, small head, and single-clawed flippers. Color is variable. Hatchlings generally have a black carapace, white plastron, and white margins on the shell and limbs. The adult...
You are exiting the U.S. Fish and Wildlife Service website
You are being directed to
We do not guarantee that the websites we link to comply with Section 508 (Accessibility Requirements) of the Rehabilitation Act. Links also do not constitute endorsement, recommendation, or favoring by the U.S. Fish and Wildlife Service.

IMAGES
VIDEO
COMMENTS
Learn what a research methodology is and how to write one for your thesis, dissertation, or research paper. Find out how to explain your methodological approach, data collection methods, and analysis method.
Learn about the definition, structure, types, and examples of research methodology for various research projects. Find out how to choose, justify, and apply the appropriate research methodology for your study.
Learn what research methodology is and how to choose the best approach for your study. Compare qualitative, quantitative and mixed methods, sampling strategies, data collection and analysis methods with examples and videos.
Learn what research methodology is, why it is important, and how to write a good one. Find out the basic structure of a research methodology and the instruments you can use to conduct your study.
Your methods for gathering data should have a clear connection to your research problem. In other words, make sure that your methods will actually address the problem. One of the most common deficiencies found in research papers is that the proposed methodology is not suitable to achieving the stated objective of your paper.
A research design is a strategy for answering your research question using empirical data. Creating a research design means making decisions about: Your overall research objectives and approach. Whether you'll rely on primary research or secondary research. Your sampling methods or criteria for selecting subjects. Your data collection methods.
Do yourself a favour and start with the end in mind. Section 1 - Introduction. As with all chapters in your dissertation or thesis, the methodology chapter should have a brief introduction. In this section, you should remind your readers what the focus of your study is, especially the research aims. As we've discussed many times on the blog ...
Learn how to choose and apply research methods for collecting and analyzing data. Compare qualitative and quantitative, primary and secondary, descriptive and experimental methods with examples and pros and cons.
Research Methodology Example. Detailed Walkthrough + Free Methodology Chapter Template. If you're working on a dissertation or thesis and are looking for an example of a research methodology chapter, you've come to the right place. In this video, we walk you through a research methodology from a dissertation that earned full distinction ...
Research methodology is the systematic process of planning, executing, and evaluating scientific investigation. It encompasses the techniques, tools, and procedures used to collect, analyze, and interpret data, ensuring the reliability and validity of research findings.
0 comment 25. Research methodology 1,2 is a structured and scientific approach used to collect, analyze, and interpret quantitative or qualitative data to answer research questions or test hypotheses. A research methodology is like a plan for carrying out research and helps keep researchers on track by limiting the scope of the research.
The first step in writing your research methodology is to explain your general approach to the research and how you will go about it. There are two ways you can do this: Option 1: Explain the ...
Learn how to write a strong methodology chapter that allows readers to evaluate the reliability and validity of the research. A good methodology chapter incl...
Here is an explanation of each step: 1. Title and Abstract. Choose a concise and descriptive title that reflects the essence of your research. Write an abstract summarizing your research question, objectives, methodology, and expected outcomes. It should provide a brief overview of your proposal. 2.
A quantitative approach and statistical analysis would give you a bigger picture. 3. Identify how your analysis answers your research questions. Relate your methodology back to your original research questions and present a proposed outcome based on your analysis.
Learn what is research methodology, how to choose the right methods for your study, and how to write a clear and effective methodology section for your research paper. This article covers the basics of quantitative, qualitative, and mixed methods, and provides a step-by-step guide based on the research onion model.
Here are eight key steps to writing a methodology: 1. Restate your thesis or research problem. The first step to writing an effective methodology requires that you restate your initial thesis. It's an important step that allows the reader to remember the most important aspects of your research and follow each step of your methodology.
The methods section of a research proposal must contain all the necessary information that will facilitate another researcher to replicate your research. The purpose of writing this section is to convince the funding agency that the methods you plan to use are sound and this is the most suitable approach to address the problem you have chosen.
Learn how to write a research proposal for your academic project, including the purpose, structure, elements, and tips. See examples of proposals for different fields and purposes.
Here are the steps to follow when writing a methodology: 1. Restate your thesis or research problem. The first part of your methodology is a restatement of the problem your research investigates. This allows your reader to follow your methodology step by step, from beginning to end. Restating your thesis also provides you an opportunity to ...
IV. Proposed Solution or Project Description: [Present your proposed solution or project in a clear and detailed manner. Explain how it addresses the problem and why it is the most effective approach. Highlight any unique features or advantages.] V. Methodology: [Describe the step-by-step approach or methodology you will use to implement your ...
INTRODUCTION. A clean, well-thought-out proposal forms the backbone for the research itself and hence becomes the most important step in the process of conduct of research.[] The objective of preparing a research proposal would be to obtain approvals from various committees including ethics committee [details under 'Research methodology II' section [Table 1] in this issue of IJA) and to ...
Content analysis methodology is used to gather and comprehend evidence in order to develop valid understandings (Seuring and Gold 2012) It is a beneficial technique to create knowledge in the area of SCM and hence performed in this research process as three stages - material selection, definition analysis and framework development and ...
The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
The percent of farm sequence-clusters with an ID change was 6.5-8.7% for our best approaches. In contrast, ~43% of farm sequence-clusters had variation in their RFLP-type, further demonstrating how our proposed fine-scale classification system addresses shortcomings of RFLP-typing.
Realizing sensitive and efficient detection of biomolecules and drug molecules is of great significance. Among the detection methods that have been proposed, electrochemical sensing is favored for its outstanding advantages such as simple operation, low cost, fast response and high sensitivity. The unique st Analytical Methods Review Articles 2024
Evaluate whether the proposed information collection is necessary for the proper functions of the Department. Evaluate the accuracy of our estimate of the time and cost burden for this proposed collection, including the validity of the methodology and assumptions used. Enhance the quality, utility, and clarity of the information to be collected.
The main heading of "Methods" should be centered, boldfaced, and capitalized. Subheadings within this section are left-aligned, boldfaced, and in title case. You can also add lower level headings within these subsections, as long as they follow APA heading styles. To structure your methods section, you can use the subheadings of ...
Numerous algorithms have been proposed to identify cell types in single-cell RNA sequencing data, yet a fundamental problem remains: determining associations between cells and phenotypes such as cancer. We develop SCIPAC, the first algorithm that quantitatively estimates the association between each cell in single-cell data and a phenotype. SCIPAC also provides a p-value for each association ...
As required by section 4(b)(2) of the Act, we use the best scientific data available to designate critical habitat. In accordance with the Act and our implementing regulations at 50 CFR 424.12(b), we review available information pertaining to the habitat requirements of the species and identify specific areas within the geographical area occupied by the species at the time of listing and any ...