

What are the Big Open Questions in Plant Cell Biology?
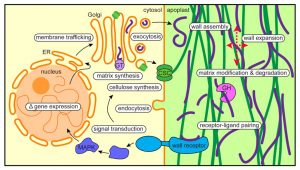
- How are organelle identity, domains, and boundaries maintained under the continuous flux of vesicle trafficking and membrane remodeling?
- Is the plant cortical microtubule cytoskeleton a mechanosensory apparatus?
- How are the cellular pathways of cell wall synthesis, assembly, modification, and integrity sensing linked in plants?
- Why do plasmodesmata open and close?
- Are there retro-signals from vacuoles to the nucleus?
- How do root cells accommodate fungal endosymbionts?
- What is the role of cell edges in plant morphogenesis?
- How is the cell division site determined?
- What are the emergent effects of polyploidy on the biology of the cell, and how are any such “rules” conditioned by cell type?
- Can mechanical forces trigger new cell fates in plants?
- How does a single differentiated somatic cell reprogram and gain pluripotency?
- How does polarity develop de-novo in isolated plant cells?
- What might the spectrum of cellular functions be for membraneless organelles and inherently disordered proteins?
- How do plants deal with internal noise?
- How does order emerge in cells and propagate to organs and organisms from chaos prone complex dynamical processes?
Some questions are age-old and have been studies for decades with great progress, yet each result opens deeper mysteries. Some questions are new, arising from a recent recognition of importance. We hope this collection of questions piques your curiosity, stimulates your interest in new topics, and maybe even generates ideas for future research. No list is ever complete. With only 15 questions, we have missed many other important mysteries. Please add your favorite open plant cell biology questions here.
Plant tissues and organs are developed from many different cells. How are these different cells cordinated and played a role under different abiotic and biotic stress conditions?
How does a cell measure its size? Interesting recent work on this question from Robert Sablowski’s group (D’Ario, M., Tavares, R., Schiessl, K., Desvoyes, B., Gutierrez, C., Howard, M., and Sablowski, R. (2021). Cell size controlled in plants using DNA content as an internal scale. Science 372, 1176–1181.)
I would like to ask a question about curriculum – what topics do you think are most important for students to be learning now, so that they will be poised to understand and take part in plant cell biology in the next decade? the reason I ask is that we are now working on the 10th edition of a cell biology textbook (Karp’s Cell and Molecular Biology) and we want to increase the representation plant cell biology. Traditionally most cell biology books like Alberts have focused almost exclusively on animal cells. In the last edition of Karp we added additional sections on plant cell biology to every chapter, but our choice of what to include was perhaps a bit arbitrary. we would love to get some guidance from the Plant Cell community!!! if you can suggest a “wish list” of topics, that would be much appreciated. please feel free to either reply in this forum or else email me directly [email protected]
Leave a Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Manage your Membership
- Join an ASPB Section
- Visit our Store
- Sponsorship & Advertising
- Make a Donation
- Read the Plant Science Today Blog
Member Services
- (301) 251-0560
Awards & Funding
- Apply for Grants & Travel Awards
Meetings & Events
- Meeting Management Services
- Plant Synthetic Biology 2021
- Plant Biology Meeting
Publications & News
- ASPB Journals
- Read The Plant Cell Blog
- Read the Plant Physiology Blog
- Submit an Article
- Read the ASPB News
- Get News & Updates
- Check out The Signal
About Plantae
- Join Plantae
- Subscribe to the Plant Science Research Weekly
- Search for Careers & Internships
- Listen to Plantae Podcasts
- Submit your Science Event to our Calendar
- *All Plantae content is licensed under a Creative Commons A-NC 2.0 License

Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Plant cell biology articles from across Nature Portfolio
Plant cell biology is the study of all aspects of plant cells. It is particularly concerned with structure, growth, division, signalling, differentiation and death of plant cells.
Related Subjects
- Chloroplasts
- Circadian rhythms in plants
- Plant cell cycle
- Plant cell death
- Plant cytoskeleton
- Plant polarity
- Plant transporters
- Protein trafficking in plants
- Proteolysis in plants
Latest Research and Reviews
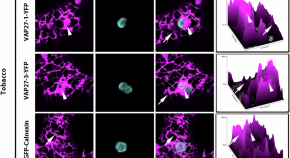
ER-associated VAP27-1 and VAP27-3 proteins functionally link the lipid-binding ORP2A at the ER-chloroplast contact sites
The plant endoplasmic reticulum (ER) interacts with other organelles at membrane contact sites. Here the authors show that plant-unique ER-chloroplasts contact sites marked by VAP27-1, VAP27-3, and ORP2a proteins function in chloroplast lipid homeostasis.
- Luciana Renna
- Giovanni Stefano
- Federica Brandizzi
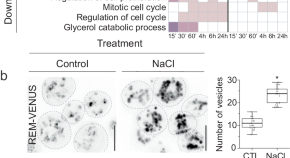
Multi-omics analysis of green lineage osmotic stress pathways unveils crucial roles of different cellular compartments
Evolution of osmoregulation allowed photosynthetic organisms to transform the whole biosphere. Leveraging high-throughput techniques in the freshwater alga Chlamydomonas reinhardtii , the authors uncover evolutionary conservation and divergence of osmoregulatory pathways within the green lineage.
- Josep Vilarrasa-Blasi
- Tamara Vellosillo
- José R. Dinneny
Evolutional heterochromatin condensation delineates chromocenter formation and retrotransposon silencing in plants
The researchers have uncovered that the phase separation ability of plant heterochromatin protein ADCP1 variants defines the formation of heterochromatic condensates (chromocenters) in different plant species.
- Weifeng Zhang
- Lingling Cheng
- Qianwen Sun
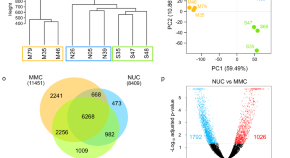
Germline β−1,3-glucan deposits are required for female gametogenesis in Arabidopsis thaliana
During female gametogenesis in plants, the Megaspore Mother Cell (MMC) accumulates β−1,3-glucan. Here Pinto et al. show that increased β−1,3-glucanase in the MMC delays β−1,3-glucan deposition, disrupts cell identity, and halts gametogenesis.
- Sara C. Pinto
- Weng Herng Leong
- Matthew R. Tucker
Distinct functions of microtubules and actin filaments in the transportation of the male germ unit in pollen
Male germ unit (MGU) transportation in pollen tubes is critical for double fertilization in flowering plants. Wang et al. reveal distinct functions of microtubule-kinesin and actomyosin system in the directional migration of MGU.
- Xiangfei Wang
Implicating the red body of Nannochloropsis in forming the recalcitrant cell wall polymer algaenan
The red body is a characteristic cellular structure of unknown function in eustigmatophyte algae. Using imaging and spectroscopy, the authors investigate the red body and propose a role in delivery of algaenan precursors for cell wall formation.
- Christopher W. Gee
- Johan Andersen-Ranberg
- Krishna K. Niyogi
News and Comment
Phragmoplast expansion requires α-Aurora
- Raphael Trösch
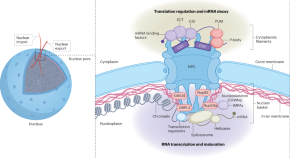
Nuclear pores beyond macromolecule channels
The nuclear pore is known as a large protein complex for the transport of macromolecules between the nucleus and the cytoplasm. Comprehensive proteomic analyses revealed a novel role of the nuclear pore complex as a platform for the coordinated regulation of the flow from transcription to translation.
- Sachihiro Matsunaga
Elmo meets Quasimodo
Interacting with both sides
- Guillaume Tena
Lignin strips in glandular trichomes
Compartmentalization of specialized compounds in glandular trichomes is essential for plant protection against stresses and attraction of pollinators. This compartmentalization is achieved by the establishment of a lignin-based apoplastic barrier named ‘neck strip’, which is present in trichomes of diverse plant species.
- Igor Cesarino
- Dyoni M. Oliveira
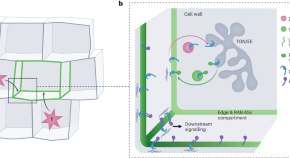
On the edge — how plant cells monitor growth
Cell edges are part of a coordinate system used by the cell to direct three-dimensional growth in an organized manner. They emerge as signalling hubs with unique composition, capable of sensing mechanical stresses derived from neighbouring cells, to negotiate and harmonize individual cellular growth rates, therefore avoiding mechanical conflict.
- Choy Kriechbaum
- Sabine Müller
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Search Menu
- Sign in through your institution
- Advance Articles
- Collections
- Focus Collections
- Teaching Tools in Plant Biology
- Browse by cover
- High-Impact Research
- Author Guidelines
- Quick and Simple Author Support
- Focus Issues Call for Papers
- Submission Site
- Open Access Options
- Self-Archiving Policy
- Why Publish with Us?
- About The Plant Cell
- About The American Society of Plant Biologists
- Editorial Board
- Advertising & Corporate Services
- Journals on Oxford Academic
- Books on Oxford Academic

High-Impact Research from The Plant Cell
Explore a collection of the most read and most cited articles making an impact in The Plant Cell published within the past two years. This collection will be continuously updated with the journal's leading articles so be sure to revisit periodically to see what is being read and cited.
Also discover the articles being discussed the most on digital media by exploring this Altmetric report pulling the articles discussed the most in the past year.

Recognizing and Celebrating Women in Science
Oxford University Press is proud to support diverse voices across our publishing. In this collection, we shine a spotlight on the representation of women in scientific fields, the gains that have been made in their fields, from research and major discoveries to advocacy and outreach, and amplify the voices of women who have made a career in scientific research.
Delve into the Women in Science collection
- Recommend to Your Librarian
- Advertising & Corporate Services
- Awards & Funding
- Plant Science Today
- Plant Biology Meeting
- Meeting Management Services
- Plant Science Research Weekly
- Taproot: A Plantae Podcast
Affiliations
- Online ISSN 1532-298X
- Print ISSN 1040-4651
- Copyright © 2024 American Society of Plant Biologists
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Help & FAQ
Top five unanswered questions in plant cell surface research
Research output : Contribution to journal › Review article › peer-review
Plant cell wall researchers were asked their view on what the major unanswered questions are in their field. This article summarises the feedback that was received from them in five questions. In this issue you can find equivalent syntheses for researchers working on bacterial, unicellular parasite and fungal systems.
| Original language | English (US) |
|---|---|
| Article number | 100121 |
| Journal | |
| Volume | 11 |
| DOIs | |
| State | Published - Jun 2024 |
All Science Journal Classification (ASJC) codes
- Microbiology
- Applied Microbiology and Biotechnology
- Molecular Biology
- Cell Biology
Access to Document
- 10.1016/j.tcsw.2024.100121
Other files and links
- Link to publication in Scopus
- Link to the citations in Scopus
T1 - Top five unanswered questions in plant cell surface research
AU - Boerjan, Wout
AU - Burlat, Vincent
AU - Cosgrove, Daniel J.
AU - Dunand, Christophe
AU - Dupree, Paul
AU - Haas, Kalina T.
AU - Ingram, Gwyneth
AU - Jamet, Elisabeth
AU - Mohnen, Debra
AU - Moussu, Steven
AU - Peaucelle, Alexis
AU - Persson, Staffan
AU - Voiniciuc, Cătălin
AU - Höfte, Herman
N1 - Publisher Copyright: © 2024 The Authors
PY - 2024/6
Y1 - 2024/6
N2 - Plant cell wall researchers were asked their view on what the major unanswered questions are in their field. This article summarises the feedback that was received from them in five questions. In this issue you can find equivalent syntheses for researchers working on bacterial, unicellular parasite and fungal systems.
AB - Plant cell wall researchers were asked their view on what the major unanswered questions are in their field. This article summarises the feedback that was received from them in five questions. In this issue you can find equivalent syntheses for researchers working on bacterial, unicellular parasite and fungal systems.
UR - http://www.scopus.com/inward/record.url?scp=85185585385&partnerID=8YFLogxK
UR - http://www.scopus.com/inward/citedby.url?scp=85185585385&partnerID=8YFLogxK
U2 - 10.1016/j.tcsw.2024.100121
DO - 10.1016/j.tcsw.2024.100121
M3 - Review article
C2 - 38405175
AN - SCOPUS:85185585385
SN - 2468-2330
JO - The Cell Surface
JF - The Cell Surface
M1 - 100121
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.34(1); 2022 Jan
A rich and bountiful harvest: Key discoveries in plant cell biology
Alice y cheung.
Department of Biochemistry and Molecular Biology, Molecular Cell Biology Program, Plant Biology Program, University of Massachusetts, Amherst, Massachusetts 01003, USA
Daniel J Cosgrove
Department of Biology, Penn State University, University Park, Pennsylvania 16802, USA
Ikuko Hara-Nishimura
Faculty of Science and Engineering, Konan University, Kobe 658-8501, Japan
Gerd Jürgens
ZMBP-Developmental Genetics, University of Tuebingen, Tuebingen 72076, Germany
Clive Lloyd
Department of Cell and Developmental Biology, John Innes Centre, Norwich NR4 7UH, UK

David G Robinson
Centre for Organismal Studies, University of Heidelberg, Heidelberg D-69120, Germany
L Andrew Staehelin
Department of Molecular, Cellular and Developmental Biology, University of Colorado, Boulder, Colorado 80309-0347, USA
Dolf Weijers
Laboratory of Biochemistry, Wageningen University, Wageningen 6708WE, the Netherlands
The field of plant cell biology has a rich history of discovery, going back to Robert Hooke’s discovery of cells themselves. The development of microscopes and preparation techniques has allowed for the visualization of subcellular structures, and the use of protein biochemistry, genetics, and molecular biology has enabled the identification of proteins and mechanisms that regulate key cellular processes. In this review, seven senior plant cell biologists reflect on the development of this research field in the past decades, including the foundational contributions that their teams have made to our rich, current insights into cell biology. Topics covered include signaling and cell morphogenesis, membrane trafficking, cytokinesis, cytoskeletal regulation, and cell wall biology. In addition, these scientists illustrate the pathways to discovery in this exciting research field.
Seven senior plant cell biologists reflect on foundational contributions to a variety of topics, including pollen tube signaling, cell morphogenesis, membrane trafficking, cytokinesis, cytoskeletal regulation, and cell wall biology.
Introduction
The concept of living organisms being composed of smaller units might have been unfathomable until Robert Hooke discovered small units, akin to monks’ cells, in cork. Since then, however, spurred by Antonie van Leeuwenhoek’s observation of living Spirogyra algae in pond water, cell theory has been central to biology. At its inception, cell biology was greatly influenced by discoveries made in plants, and plant cell biology has since been an active research field that has given birth to many exciting concepts and discoveries. Starting in the middle of the 20th century, new tools and methods have allowed researchers to explore sub-cellular processes and take “inventory of our acquired wealth,” as stated by Albert Claude in his Nobel Lecture in 1974. Plant cells not only share many features with the eukaryotic cells studied by Claude and others, but also have distinct structures and processes. In fact, new cellular structures and organelles are still being identified today, and we can rest assured that plant cell biology will continue to reveal the plant’s inner secrets. Given the wealth of discoveries on which our current textbooks are built, it would be unrealistic to exhaustively cover the major findings and highlight mechanisms and concepts across this broad field. Instead, in this review, seven senior plant cell biologists offer their personal reflections on the discoveries they witnessed in their respective domains within the field of plant cell biology. While these are personal retrospectives, these accounts illustrate what we did not know, what we have learned, and to some extent which questions still beg to answer.
Alice Cheung: Journeying with the pollen tube to finding a versatile signaling module for reproduction and beyond
Alice Cheung describes how, over the course of her career, her work has explored the nature of how cells grow at their tips, starting with pollen tubes and later including root hairs. Key findings are how small GTPases and actin-regulatory proteins coordinate the action of the actin cytoskeleton to support tip-focused growth. These works led to the identification of a cell-surface signaling module with broad functional implications for plant growth, reproduction, and coping with environmental challenges.
Lured by the inner beauty of a pollen tube, I entered the field of plant cell biology when live-cell imaging was becoming mainstream. Pollen tubes grow exclusively at their tips to deliver sperm for fertilization and have been a classical system for studying polarized growth. Growing pollen tubes maintain a dynamic and highly polarized cytoplasmic organization, with a mesh-like actin structure at the subapical region subtending an apical collection of vesicles and long actin cables in the distal organelle-rich cytoplasm ( Figure 1A ). Studies spanning decades performed by a large community of biologists have contributed to our understanding of the processes through which cell membrane and wall materials are carried to the elongating tip in vesicles that move along these actin cables ( Cheung and Wu, 2008 ).

From the pollen tube to a cell surface signaling module with functions in diverse processes. A, Hallmark features in an elongating pollen tube: an apical vesicular zone (red), subapical actin (green/yellow) mesh, and long actin cables. B, Formin-regulated actin assembly. (Left) FORMIN HOMOLOGY PROTEIN1-stimulated assembly. Image shows three contiguous 1 µm optical sections showing bundles of actin (arrowhead) emanating from the cell membrane (top to bottom). (Middle and right) FORMIN HOMOLOGY PROTEIN5-generated subapical actin mesh (arrowheads). The middle panel shows the assembly of a subapical actin mesh in a pollen tube as it emerged from growth arrest to rapid polarized growth; arrowheads, nascent short actin bundles. The right panel highlights an annular configuration of the subapical actin structure (arrows). C and D, Protein pull-down assays showing that pollen hydration (C) and auxin (D) activate RAC/ROPs (upper panels). E, Nuclei of plant cells expressing IAA17-GFP showing rapid formation of nuclear protein bodies (arrow, as an example) in response to auxin. F, The FERONIA–LLG–RALF trimeric signaling module. G, Wild-type, feronia , and llg1 plants showing the impacts of loss of the signaling module on growth and development. H–J, Functions of FER–LLG1–RALF-related signaling modules in reproduction. H, FERONIA-LLG1-RALF controls stigma ROS to regulate pollen germination. I, Pollen tube growth/integrity; image shows that only half the ovules were fertilized (reflected by blue dots) by pollen tubes from a heterozygous male parent (BUPS1/-). J, FERONIA controls multiple steps in pollen–pistil interactions: in wild-type pistils, single pollen tubes exit from the septum (arrows) and enter individual ovules to release sperm for fertilization. In feronia pistils, bundled pollen tubes exit from the septum (arrowheads) and enter into individual ovules; asterisks show pollen tube pile-up from nonruptured pollen tubes in mutant female gametophytes. Scale bars: 10 µm (A and B), 5 µm (E), 3 cm (F), 250 µm (I), and 100 µm (J). A from Cheung and Wu (2004) Figure 6D; (B) adapted from Cheung and Wu (2004) Figure 4A and Cheung et al. (2010) Supplemental Figure S5; (C) reprinted from Chen et al. (2013) Figure 1A , with permission from Elsevier; (D) from Tao et al. (2002) Figure 5C; (E) from Tao et al. (2005) Figure 5, A and B ; (F) modified from Duan et al. (2010) Figure 3K and Li et al. (2015) Figure 11 ; (G) modified from Li et al. (2015) Figure 1C; (H) Adapted from Zhang et al., 2021a Figure 7A ; (I) from Ge et al. (2017) Supplemental Figure S6B, reprinted with permission from AAAS; (J) from Duan et al. (2020) Figure 1B .
Our early cell biological studies helped establish small GTPases of the RHO (RAC/ROPs) and RAB families as important regulators of actin dynamics and vesicular trafficking in pollen tubes (Chen et al., 2002 , 2003 ; Cheung et al., 2002 ; de Graaf et al., 2005 ). We pioneered the study of formins (actin-nucleating proteins that stimulate the assembly of linear actin) in plant cells and demonstrated the role of nascent actin assembly for pollen tube growth ( Figure 1B; Cheung and Wu, 2004 ; Cheung et al., 2010 ). These excursions added a cell biological context to our already ongoing molecular and biochemical work in male–female interactions ( Wang et al., 1993 ; Cheung et al., 1995 ; Wu et al. 1995 ). We later incorporated root hairs, another tip-growing system, and seedlings into our studies. This merger has worked well because the seedling system allowed us to carry out fine mechanistic dissections, generating findings that provided hints to explore the considerably less accessible reproductive processes. Together, they led to the discoveries that pollen grain hydration ( Figure 1C ) and auxin treatment of seedlings ( Figure 1D ) both rapidly activate RAC/ROP GTPases ( Tao et al., 2002 ; Chen, 2003 , 2013 ; Wu et al., 2011 ). We also observed that auxin and RAC/ROPs mediate the assembly of nuclear protein bodies that comprise AUX/IAA repressors and components of the ubiquitin/26S proteasome apparatus, and they self-degrade the AUX/IAA repressors in response to auxin ( Figure 1E ). In hindsight, and given the emerging knowledge about ubiquitylation-dependent phase separation of the proteasome (e.g. Yasuda et al., 2018 ), it would be interesting to examine whether the auxin–RAC/ROP- and substrate-mediated processes for nuclear protein body formation are a manifestation of liquid–liquid phase transition. Given that RAC/ROPs are the major molecular switches that act along the inner cell membrane, we turned to searching for upstream regulators of RAC/ROPs to link signal perception to signal transduction.
Starting from using ROP-GEF1, a guanine nucleotide exchange factor that activates RAC/ROPs, as bait in yeast two-hybrid assays, we discovered the FERONIA receptor kinase as a surface regulator for RAC/ROPs ( Duan et al., 2010 ). FERONIA functions with the glycosylphosphatidylinositol-anchored protein LLG1 ( Li et al., 2015) as co-receptors on the cell surface ( Figure 1F ). FERONIA-LLG1 activates RAC/ROP-mediated reactive oxygen species (ROS) production by NADPH oxidase and intersects with multiple pathways to broadly affect growth and development. In reproduction, FERONIA and LORELEI, the ovule-specific LLG1 homolog, turn out to be critical for female fertility, since the FERONIA/LORELEI-to-ROS pathway is required for pollen tube bursting to release sperm in the female gametophyte, enabling fertilization ( Figure 1J; Duan et al., 2014 ). We also showed that variants of the FERONIA–LORELEI coreceptor and ROS are important throughout pollination, controlling pollen germination on the stigma through RAC/ROP-mediated ROS production ( Liu et al. 2021 ; Zhang et al., 2021a , 2021b ) and preventing premature pollen tube bursting during the growth process ( Ge et al., 2017 , 2019 ; Figure 1, H and I ).
FERONIA is a member of the malectin-like domain-containing receptor-like kinase family ( Cheung and Wu, 2011 ; Doblas et al., 2018 ; Yang et al., 2021 ). A pioneering study of THESEUS1, which is closely related to FERONIA, showed that it functions as a sensor of cell wall perturbations to suppress the growth of cellulose-deficient seedlings ( Hematy et al., 2007 ). We established biochemically that the extracellular domain of FERONIA binds pectin, a major cell wall polysaccharide ( Feng et al., 2018) . This property affects the pectic environment along the pollen tube growth pathway and affects how plants avert polyspermy ( Duan et al., 2020 ; Figure 1J ). The crucial roles of FERONIA in growth and development ( Figure 1G; Li et al., 2016 ), and discoveries from Michael Sussman’s and Cyril Zipfel’s laboratories that peptide regulators known as RAPID ALKALINIZATION FACTORS (RALFs) are ligands for FERONIA that regulate growth and immunity ( Haruta et al., 2014 ; Stegmann et al., 2017 ) considerably expanded the field beyond reproduction. RALFs that regulate FERONIA and related receptor kinase-controlled processes in female tissues or pollen have also been identified ( Ge et al., 2017 , 2019 ; Liu et al., 2021) .
How FERONIA–LLG1/LORELEI achieves its diverse functions remains unclear. Sensing multiple peptides as ligands, partnering with different LLG homologs, and utilizing the broadly functional RAC/ROPs and ubiquitous ROS as signaling mediators can all result in functional diversification. In seedlings, the FERONIA–pectin interaction has been linked to cell wall integrity; the loss of FERONIA renders seedlings more prone to salinity-induced cell bursting ( Feng et al., 2018) . The cell wall encounters myriad endogenous and extracellular signals from neighboring cells and the environment. The ability of FERONIA to interact with the cell wall could also contribute to its diverse biological roles. Sorting out how the FERONIA–LLG1 signaling module achieves its broad and profound functions in growth and survival will be a formidable challenge to tackle.
While working on two developmentally distant systems sometimes appeared chaotic, it has turned out to be immensely rewarding. Following logical next steps, even when it meant taking time to start from scratch so we could venture into unfamiliar territory, has helped us build perspectives. Discovering FERONIA as an upstream regulator of RAC/ROPs in seedlings and that FERONIA turns out to be a major regulator of female fertility ( Escobar-Restrepo et al., 2007 ) was a lucky coincidence. It brought us back from pollen tube cell biology to where we started in male–female interactions and stimulated many exciting findings from many groups. The Plant Cell is home to many of the studies cited here. Elucidating the huge signaling capacity harnessed in large numbers of potential variants of the FERONIA–LLG–RALF signaling complex assembled from related proteins ( Yang et al., 2021 ), their mechanisms, and their molecular interactions at high resolution will likely remain rich grounds for contributions to The Plant Cell in years to come.
Daniel J. Cosgrove: Stress relaxation, expansins, and cell wall structure
Daniel Cosgrove’s work has focused on understanding the complex biology and biochemistry that underlies the properties of the growing cell wall. A major focus of his work has been toward characterizing the contribution of cell wall loosening to cell growth. His career spans from classical physiological studies, through molecular identification of the expansin protein family, to modern modeling approaches.
Irreversible cell enlargement (growth) requires concomitant water uptake and wall yielding. Decades of conflicting opinions about how these two physically distinct processes are causally connected went unresolved until Ray et al. (1972) clarified that wall “stress relaxation is the primary event in cell enlargement, whereas water uptake, volume increase, and extension (strain) of the cell wall are secondary.” They noted that “explicit treatment of the stress relaxation process…seems not to have been given in the plant growth literature.” Although stress relaxation was measured empirically in isolated walls ( Yamamoto et al., 1970 ; Cleland and Haughton, 1971 ), how it instigated water uptake and growth in vivo was clarified only later by a new theory combined with experimental methods to measure stress relaxation in vivo ( Cosgrove, 1985 , 1987a ). The principle of the method was to prevent cell water uptake and to measure the ensuing decay in turgor pressure as the wall relaxed. This work, along with related studies, provided the framework to determine which biophysical parameters control cell growth. In theory, either water uptake or wall yielding could limit cell growth, or the two processes could co-limit growth. A review of this field concluded that “reduction in cell turgor pressure, as a consequence of wall relaxation, serves as the major initiator and control point for plant cell enlargement” ( Cosgrove, 1993 ).
Perhaps, you have seen the cartoon of a theoretician scrawling equations on a blackboard, leading to the punchline “Then a miracle occurs!” To some biologists, stress relaxation may seem a bit like that. To put a molecular face on wall stress relaxation, we may ask: what is it, what causes it, and how is it controlled? Physically, wall stress relaxation is a reduction in the tensile forces transmitted between polymers in the wall. This may originate with the cutting of load-bearing tethers within the wall, viscoelastic movements of wall polymers toward a lower energy state, or the release of noncovalent bonding between wall components. A satisfying answer to this question calls for a molecular model of the cell wall that includes the dynamic interactions of wall polysaccharides and their responses to forces, that is, a molecular model of wall mechanics.
The discovery of expansins opened one avenue to investigating this “miracle.” For many years, acid pH was known to stimulate wall expansion (dubbed “acid growth”), leading to the speculation that pH-sensitive hydrolases cut load-bearing crosslinks in the cell wall. Acid growth is best known in connection with auxin-stimulated growth ( Rayle and Cleland, 1992 ; Du et al., 2020 ), but it is also connected to many other stimuli that modulate growth. A concerted hunt for the catalysts behind acid growth led us to discover α-expansins (EXPA; McQueen-Mason et al., 1992 ). These small proteins are present in growing cell walls in catalytic amounts and their activity is maximal at low pH (∼4) and minimal at neutral pH. The “trick” in identifying EXPAs was the use of a constant-force extensometer to measure cell wall creep (time-dependent, irreversible extension). Acid-induced creep in native cell walls was known to require endogenous proteins, potentially enzymes ( Cosgrove, 1989 ). Contrary to the enzyme hypothesis, however, EXPAs lack enzymatic activity ( Cosgrove, 2020 ). They rapidly induce wall creep without weakening the cell wall ( McQueen-Mason and Cosgrove, 1995 ; Yuan et al., 2001 ).
After the cloning of EXPA ( Shcherban et al., 1995 ), it became apparent that expansins in land plants comprise a multigene superfamily, consisting of two moderately large families, EXPAs and β-expansins (EXPBs), and two smaller groups (EXLA and EXLB; Sampedro and Cosgrove, 2005 ). Differential expression of EXPA genes is connected with cell growth and many other developmental processes involving wall loosening ( Figure 2 ). The role of EXPBs has not been studied in depth, except for an evolutionary outlier, a group of proteins expressed abundantly and specifically in grass pollen ( Sampedro et al., 2015 ). Unlike EXPAs, these pollen EXPBs solubilize the dominant hemicellulose in grass cell walls, arabinoxylans, but without lytic activity ( Yennawar et al., 2006 ; Tabuchi et al., 2011 ; Wang et al., 2016 ). This action promotes penetration of growing pollen tubes between stylar cells ( Valdivia et al., 2009 ). Much remains to be discovered about the physical actions and biological roles of the other EXPBs.

Evolution of expansin-related research topics. The figure depicts the development of the expansin research field, starting with the discovery of EXPA as the mediator of acid growth to its involvement in many developmental processes, the discovery of EXPBs with a special function in grass pollination biology, the evolution of expansins as a multigene family, and the discovery of bacterial expansins by way of protein crystallography. Image copyrigth Daniel J. Cosgrove, used with permission.
When the structure of the maize ( Zea mays ) pollen EXPB ZmEXPB1 was solved by X-ray crystallography ( Yennawar et al., 2006 ), we learned it consists of two tightly packed domains with a long, flat surface suitable for polysaccharide binding. The N-terminal domain is homologous to family-45 endoglucanase, but it lacks the catalytic base normally needed for enzymatic activity. The C-terminal domain has a structure suggestive of a carbohydrate-binding domain. This structure led to the discovery of a structurally homologous protein from the bacterium Bacillus subtilis ( Kerff et al., 2008) and thereafter the recognition that many phylogenetically diverse microbes encode expansins in their genomes, with many of these proteins likely involved in plant–microbe interactions ( Nikolaidis et al., 2014 ; Narvaez-Barragan et al., 2020 ).
Although the structures of expansins are known in atomistic detail, how they induce wall stress relaxation and creep remains uncertain. Recent results from xyloglucanase digestions ( Park and Cosgrove, 2012 ; Zhang et al., 2019 ) cast doubt on traditional depictions of cell walls in which cellulose microfibrils are connected together via xyloglucan or pectin tethers. Consequently, we recently took a fresh approach by combining coarse-grained modeling with mechanical assays to test the physical soundness of cell wall models ( 2021b ). Single chains of cellulose, xyloglucan, and pectins were simulated with coarse-grained bead-and-spring models parameterized to correspond to the different physical properties of these wall polysaccharides. Multi-lamella walls were constructed from these model polymers, stretched in silico, and the stress–strain behaviors compared with real walls. The results: cellulose microfibrils spontaneously assembled into 2D networks that carried most of the (in-plane) tensile force, whereas pectin and xyloglucan transmitted a tiny proportion of the tensile force. Wall elasticity arose from straightening of the cellulose microfibrils, while plasticity originated from sliding between laterally bonded cellulose microfibrils. This model provided insights into the molecular bases of wall mechanics, but did not include the loosening action of expansins—a challenge for future research.
Ikuko Hara-Nishimura: Endomembrane dynamics-based physiological functions of the endoplasmic reticulum and vacuoles
Ikuko Hara-Nishimura’s work has focused on the dynamics of the endoplasmic reticulum (ER) and vacuole. Although the textbooks tell us that the ER passes proteins to the Golgi, in truth there are many different fates for proteins exiting the ER, some developmentally regulated and some stress responsive. Her work has identified functions for numerous specialized endomembrane compartments, and many regulators of their dynamics.
Here, I describe some of our discoveries of the endomembrane dynamics-based roles of the vacuole and ER. Before the era of the model plant Arabidopsis thaliana , vacuolar protein transport was investigated using developing seeds of a variety of plants, in which large amounts of storage proteins are synthesized on the ER and then transported to protein storage vacuoles. Analysis of pulse-chased developing pumpkin ( Cucurbita sp.) seeds led us to discover a Golgi-independent pathway ( Hara-Nishimura et al., 1985 ) in which proprotein precursors are delivered from the ER to the vacuoles via precursor-accumulating vesicles (PAC vesicles) ( Figure 3A ). PAC vesicles have >300 times the volume of COPII vesicles and enable efficient mass protein transport. They are generated under the stressed condition of excessive protein synthesis on the ER (discussed later). A membrane protein of PAC vesicles was later demonstrated to be a vacuolar sorting receptor (VSR; Shimada et al., 2002 , 2003 ; Figure 3A ).

Endomembrane dynamics of the ER and vacuoles to support a variety of physiological functions. A, Ultrastructure of developing pumpkin seed cells showing PAC vesicles (arrows) responsible for Golgi-independent transport of storage protein precursors to protein storage vacuoles. VSR was discovered from isolated PAC vesicles. VPE converts proproteins to functional mature proteins in the vacuoles. B, GFP-labeled ER network of an A. thaliana cotyledon cell. ER strands rapidly stream along the longitudinal axis of the cell. The ER body is a large ER-derived organelle required for single-cell chemical defense in Brassicaceae plants. C, Endomembrane dynamics responsible for various physiological roles. The ER has the ability to generate distinct compartments with specialized functions. Vacuoles have the ability to remodel their membranes for defense against pathogen infection.
Immediately after reaching the vacuole, proprotein precursors are converted into mature proteins. We biochemically isolated an enzyme responsible for the conversion from developing castor bean ( Ricinus communis ) and designated it vacuolar processing enzyme (VPE; Hara-Nishimura et al., 1993 ; Figure 3A ). VPEs play various roles in the plant lifecycle including programmed cell death ( Yamada et al., 2020a ). We subsequently established VPE/AEP -knockout mouse and demonstrated that VPE/AEP is required for the maturation of lysosomal proteins ( Shirahama-Noda et al., 2003 ). This is one of the rare cases in which plant studies provided insights into animal mechanisms. Many discoveries from A. thaliana studies have since contributed to our understanding of human biology and health ( Jones et al., 2008 ).
We were fascinated with vacuolar membrane dynamics and discovered their association with defense strategies ( Shimada et al., 2018a ; Figure 3C ). Viral infection triggers the vacuolar membrane to collapse in a VPE-dependent manner, resulting in the release of hydrolytic enzymes that prevent viral propagation in the cytosol ( Hatsugai et al., 2004 ). On the other hand, bacterial infection causes the vacuolar membrane to fuse with the plasma membrane, resulting in the release of defense proteins that prevent bacteria from proliferating in extracellular spaces ( Hatsugai et al., 2009 ). The discovery of these vacuolar protein relocations taught us that vacuoles are not necessarily the final destination or workplace of “vacuolar proteins.
The explosion of information about the model plant A. thaliana allowed us to significantly extend our knowledge about the functional peptides that are synthesized on the ER and then secreted to be used for the differentiation of target cells ( Figure 3C ). By focusing on A. thaliana genes encoding small proteins with signal peptides, we identified the peptide hormone stomagen, which positively regulates stomatal density in leaves, thereby increasing the efficiency of CO 2 uptake ( Sugano et al., 2010 ).
The development of imaging techniques has greatly extended our understanding of vesicle trafficking in plant cells (see also “Robinson’s” and “Staehelin’s” sections). A recent study combining variable-angle epifluorescence microscopy with high-temporal resolution imaging prompted us to develop a dynamic capture-and-release model of ER exit sites (ERESs) by Golgi stacks ( Takagi et al., 2020 ). This model suggests that punctate ERESs wandering into an ER network cavity are captured by a Golgi stack for cargo transfer and are then released to reload with cargo and repeat the transfer ( Figure 3C ).
Furthermore, live cell imaging showed us that plant-specific myosin XI motors play critical roles in actin-myosin cytoskeleton-dependent dynamics of the endomembranes (see also “Cheung’s” section). We established multiple mutants of myosin XIs and elucidated their functions. First, myosin XI-I links the nuclear membrane to actin filaments and moves the nucleus in response to environmental stimuli ( Tamura et al., 2013 ). Second, three myosin XIs (XI-K, XI-1, and XI-2) drive extensive streaming of the ER ( Ueda et al., 2010 ; Figure 3B ). Considering the fact that the ER has the largest endomembrane surface area in the cell, ER streaming might be the force driving cytoplasmic streaming, which has been a mystery since the late 18th century. Third, two myosin XIs (XI-F and XI-K) in elongating cells play roles in vigorous cytoplasmic streaming and organ straightening to adjust plant posture ( Okamoto et al., 2015 ). These findings seem to reveal some linkage between endomembrane dynamics and organ movements.
Finally, I would like to emphasize that the ER has the ability to generate specialized compartments in response to stress ( Figure 3C ). As described above, PAC vesicles are generated from the ER in response to the abundant production of storage proteins ( Figure 3A ). Similarly, the abundant production of KDEL-ER-retention signal-tailed cysteine proteinases triggers the generation of specific organelles from the ER ( Schmid et al., 1998 ; Okamoto et al., 2003 ). The largest ER-derived compartments in Brassicaceae plants are ER bodies ( Figure 3, B and C ). ER bodies establish a single-cell chemical defense in which damage from herbivore feeding causes ER-body β-glucosidases to react with glucosinolates and release repellent compounds ( Yamada et al., 2020b ). The question raised is how these organelles are generated from the ER in response to stress. We have recently succeeded in generating ER bodies in nonBrassicaceae plants including monocots by expressing two Brassicaceae-specific genes ( NAI2 and BGLU23 ) ( Yamada et al., 2020b ). This discovery could be used to produce herbivore-resistant crops.
Another question is how the sizes of ER-derived organelles are determined. An answer was provided from studies of oil bodies that are generated from the ER ( Figure 3, A and C ). Membrane proteins function in maintaining the proper sizes of oil bodies in seed cells, which preserves the viability of over-wintering oilseeds ( Shimada et al., 2018b ). We discovered two roles for oil bodies in leaves. First, oil bodies in fungal-infected and senescent leaves function as subcellular factories to produce antifungal compounds ( Shimada et al., 2018b ). Second, oil bodies that accumulate sterol esters (SEs bodies) are generated in response to the excess production of sterols ( Shimada et al., 2019) . SE bodies function in sterol homeostasis, in which excess sterols are converted to nontoxic sterol esters on the ER subdomains, which develop into SE bodies.
In conclusion, the ER and vacuole are flexible and dynamic endomembrane organelles. They and their membranes are remodeled in response to internal and external stresses. The ER has the ability to generate distinct compartments in response to the overproduction of materials, while vacuoles have the ability to remodel their membranes in response to pathogen infection. Their characteristic features might contribute to organelle engineering to create valuable crops.
Gerd Jürgens: Membrane fusion in plant cytokinesis
Gerd Jürgens entered plant cell biology through studying Arabidopsis embryogenesis, starting from genetic screens for patterning mutants. In addition to several key regulators of cell polarity and cell fate, he identified a wide range of genes involved in conserved cellular processes. Here he describes how this work uncovered key components of the machinery involved in membrane fusion, both during cytokinesis and in the secretory pathway.
Cytokinesis marks the transition from one cell generation to the next by physically partitioning the dividing cell into two daughter cells. Although cytokinesis is a hallmark of all living systems, land plants (and most streptophytic algae) have evolved their own strategy—phragmoplast-assisted cell-plate formation—which contrasts with constriction from the surface, a mechanism shared by nonplant organisms with or without cell walls. Cell-plate formation was first described at the ultrastructural level in the 1960s ( Hepler and Newcomb, 1967 ). However, modern functional analysis only started in the 1990s, with Arabidopsis playing a prominent role.
The mutational beginning
Our early research focused on the genetic analysis of embryo pattern formation. Among the many mutants isolated in a large-scale screen, we identified several alleles of two genes named KNOLLE ( KN ) (German for “tuber”) and KEULE ( KEU ) (German for “club”) that proved to play key roles in cell-plate formation ( Mayer et al., 1991 ). In those days, getting hold of the relevant DNA sequence was a real feat: since there was no genome sequence to lean on, this analysis involved chromosome walking with yeast artificial chromosome (YAC) libraries and mapping of (hopefully) polymorphic YAC ends by genetic recombination relative to the mutation of interest. Eventually, the two genes were isolated and shown to encode a “syntaxin-related gene product” and “a SEC1 protein that binds the syntaxin KNOLLE,” respectively ( Lukowitz et al., 1996 ; Assaad et al., 2001 ; Figure 4 ). The significance of these findings was not immediately clear to us, although the SNARE (short hand for soluble N-ethylmaleimide-sensitive-factor attachment receptor) hypothesis had just been formulated on the basis of biochemical studies in mammals and molecular analysis of yeast genes involved in protein secretion ( Söllner et al., 1993 ; Ferro-Novick and Jahn, 1994 ). SNAREs are integral membrane proteins that form stable complexes of three or four SNARE proteins. These complexes contribute to the selective fusion of juxtaposed (endo)membranes such as membrane vesicles and their target membranes (heterotypic fusion) or two equivalent membranes (homotypic fusion; Jahn and Scheller, 2006 ). Different SNARE complexes are engaged in different membrane fusions, for example, the fusion of COPI vesicles with the ER or AP1 vesicles with the plasma membrane. Each SNARE complex comprises cognate members of different subfamilies named Qa, Qb, Qc, or R-SNAREs according to the amino acid residue in the center of their SNARE domain. In addition, the so-called Qbc-SNAREs have two SNARE domains. SNARE complexes comprise Qa, Qb, Qc, and R SNAREs or Qa, Qbc, and R-SNAREs. More importantly to us, the cell cycle-regulated accumulation of KNOLLE mRNA and protein, as well as the discovery of “unfused vesicles accumulating in the plane of cell division,” in knolle and keule mutant embryos and the absence of cytokinesis altogether in knolle keule double-mutant embryos hinted at the proteins’ roles in cell-plate formation ( Lauber et al., 1997 ; Waizenegger et al., 2000 ).

Membrane fusion during cytokinesis (model). A, Membrane vesicles arriving in the plane of cell division contain cis-SNARE complexes on their surfaces. Following disassembly, presumably by the AAA ATPase NSF and its co-factor α-SNAP, the now monomeric Qa-SNARE and R-SNARE proteins are captured by SEC1-related KEULE, which not only prevents reformation of the cis-SNARE complexes but also promotes trans -SNARE complex formation and thus the fusion of vesicles with each other and with the margin of the growing cell plate. Only complexes involving Qa-SNARE KNOLLE (green) and Qbc-SNARE SNAP33 (yellow) are depicted (see B, upper right). B, Evolutionary change of Qa-SNAREs involved in cytokinesis. The ancient SNARE complexes involving Qa-SNARE SYP132 appear conserved throughout the Streptophyta, mediating vesicle fusion during both secretion and cytokinesis. Cytokinesis-specific Qa-SNARE KNOLLE originated by gene duplication with the advent of flowering plants. The encoded protein retained the interaction with the SNARE partners of SYP132, forming two types of cytokinesis-specific SNARE complexes.
The boon and bane of genetic redundancy
Genetic analysis is a powerful approach for dissecting complex biological processes. In practice, however, related genes (paralogs) often functionally replace each other and thus mask deleterious mutational effects. Both knolle and keule mutant embryos gave rise to (abnormal) seedlings, although vesicle fusion during cytokinesis was disrupted from early on. The other source of frustration was our inability to identify KNOLLE-interacting SNARE partners based on knolle -like mutant phenotypes. A screen for yeast two-hybrid interactors pulled out Qbc-SNARE SNAP33. However, its knockout mutant only had a very mild cytokinesis defect and unrelated symptoms of cell death ( Heese et al., 2001 ). At the same time, Qb-SNARE NPSN11 was identified as another KNOLLE interactor, and its knockout mutant was even viable ( Zheng et al., 2002 ). The SNARE complexes known from yeast and mammals suggested that these two KNOLLE interactors might belong to different SNARE complexes or that the SNARE complex involved in cytokinesis might have an unusual composition. Eventually, this problem was solved. The snap33 npsn11 double mutant displayed a strong knolle -like phenotype, including the accumulation of unfused vesicles in the division plane, and co-immunoprecipitation analysis revealed two KNOLLE-containing SNARE complexes, one comprising SNAP33 and the other NPSN11 ( El Kasmi et al., 2013 ). Why KNOLLE forms two different SNARE complexes that both contribute to cell-plate formation is a mystery.
The enlightening evolutionary perspective
KNOLLE redundancy was more difficult to unravel. A survey of SYP1 Qa-SNAREs revealed that SYP132 accumulated in all tissues and, together with KNOLLE, was the only SYP1 expressed in embryos ( Enami et al., 2009 ). Moreover, the knolle mutant was rescued by expression of SYP132, but not SYP121 (aka PEN1), from the KNOLLE cis-regulatory sequences ( Reichardt et al., 2011 ). Thus, SYP132 was the top candidate for a Qa-SNARE that shares a redundant function with KNOLLE in cytokinesis. This notion was further backed by the following evolutionary perspective: Cell division via cell-plate formation only arose in the streptophytic algae, while KNOLLE originated much later with the transition to flowering plants. The genome sequence of the filamentous charophyte green alga Klebsormidium nitens (aka K. flaccidum ), which divides without cell-plate formation, had just become available ( Hori et al., 2014) . We identified single-copy genes encoding orthologs of SYP132 and SNARE partners of KNOLLE (SNAP33, NPSN11, SYP71, and VAMP721). Their Arabidopsis counterparts indeed formed complexes, and syp132 knolle embryos were multinucleate, resembling knolle keule double-mutant embryos ( Park et al., 2018) . The ancient SYP132 complexes were apparently supplemented with the KNOLLE complexes in flowering plants, most probably to meet the higher demand for membrane fusion in endosperm cellularization—a unique process associated with double fertilization in flowering plants.
The KNOLLE interactor KEULE plays a central role in trans-SNARE complex formation, mediating the fusion of membrane vesicles during cell-plate formation ( Park et al., 2012 ). How KEULE might do so can be inferred from structural analysis of SEC1-related Vps33p, which functions in yeast vacuolar fusion, a topologically comparable process ( Baker et al., 2015 ). KEULE also has counterparts with redundant functions that only originated during the evolution of flowering plants. Here, the paralog SEC1B plays a major role in secretion, leaving the lion’s share of the role in cytokinesis to KEULE, which is convincingly demonstrated in the cytokinesis phenotypes of qa-snare sec1 double-mutant embryos ( Karnahl et al., 2018 ).
Other knowns and unknowns of membrane trafficking in cytokinesis
Cell-plate formation is a secretory process, with contributions from endocytosis ( Reichardt et al., 2007 ). However, trafficking from the trans-Golgi network (TGN) to the division plane is entirely regulated by the late-secretory ADP-ribosylation factor guanine-nucleotide exchange factors (ARF-GEFs) BIG1-4 ( Richter et al., 2014) . Blocking secretory trafficking to the plane of cell division at different stations along the route revealed the formation of cis-SNARE complexes in the ER, which are only disassembled upon arrival at the division plane to form trans-SNARE complexes required for cell-plate formation ( Karnahl et al., 2017 ). This disassembly requires N-ethylmaleimide-sensitive factor (NSF) ATPase activity which, however, has not been identified. The membrane vesicles that generate the cell plate pose another problem—are they homogeneous or are there KNOLLE-bearing and SYP132-bearing subpopulations? To answer this question, it might be necessary to isolate cytokinetic vesicles and determine their cargo—another daunting task.
Clive Lloyd: Early MAPs—constructing dynamic microtubule arrays
Clive Lloyd’s work has focused on the dynamic network of microtubules that mediate cellular processes. These dynamic elements of the cytoskeleton are constantly assembling and disassembling, and have crucial roles in mitosis. Microtubules are also reorient to direct plant growth and cell wall deposition. Here he describes the work of his group and others in identifying plant-specific microtubule-associated proteins (MAPs) that function to coordinate microtubule dynamics and cellular morphology.
It is hard to think back to a time when microtubules were not dynamic. Electron microscopy studies on fixed, embedded cells had established that parallel groups of cortical microtubules co-aligned with cellulose microfibrils upon the plasma membrane. These microtubules could be in transverse, oblique, or longitudinal arrays that, with the onset of mitosis, bunched into a transient preprophase band (PPB), which forecast the alignment of the new cross-wall. It was inferred from these static images that cortical microtubules must reorientate—as was suspected when cells change the direction of expansion—but nothing was known about the mechanics nor of the MAPs that regulated behavior. Our only models were high and low molecular weight protein bands (e.g. MAP2 and tau) that co-purified with brain microtubules through cycles of assembly/disassembly (but let us not forget that this stringent method failed to detect the kinesin motor protein superfamily).
In the 1970s, I was investigating the cytoskeleton in animal cells and, when given the opportunity of forming a lab working on plant cells, decided on a dual approach: trying to identify plant-specific MAPs and applying immunofluorescence microscopy, which had already transformed the animal field. The significance of the latter approach was that it introduced a change in scale by allowing microtubule arrays to be seen in their unsectioned entirety ( Lloyd et al., 1979 ). Then, in 1984, animal cell biology was revolutionized by studies using microinjected fluorescent tubulin showing that microtubules could dynamically interconvert between growing and shrinking states ( Mitchison and Kirschner, 1984 ). By microinjecting neurotubulin into Tradescantia hairs, Peter Hepler’s group found that microtubules in immobile plant cells were, paradoxically, even more dynamic than in motile animal cells ( Hush et al., 1994 ). And by microinjecting fluorescent brain tubulin into pea ( Pisum sativum ) epidermal cells, lab member Ming Yuan established that dynamicity extended to the whole cortical array when he saw cortical microtubules reorient through 90° in quite an unexpected way ( Yuan et al., 1994 ).
In turn, microinjection was supplanted by green fluorescent protein (GFP) technology and, by using long-term time-lapse imaging of fluorescent fusion proteins, Jordi Chan ( Chan et al., 2003 ; Lloyd and Chan, 2004 ) revealed how microtubule mosaics rotate around the outer face of Arabidopsis cells ( Chan et al., 2007 ). Because microtubules guide the movement of cellulose synthase complexes along the plasma membrane ( Paradez et al., 2006 ), microtubule rotation should also cause the synthase tracks to rotate, with implications for the layering of cellulose microfibrils in the cell wall. We confirmed that GFP-cellulose synthase 3 tracks also rotate, like the hands of a clock, and that this requires dynamic microtubules ( Chan et al., 2010 ). These emergent properties of microtubule behavior—which cannot be intuited from reading the genome—are crucial to plant development. Adrian Sambade demonstrated this by selecting light-sensitive mutants in the DELLA/gibberellin signaling pathway and finding that light affects microtubule dynamics and their ability to switch growth axes ( Sambade et al., 2012 ).
As far back as 1980, my colleague Toni Slabas demonstrated that presumptive but unidentified plant MAPs could be isolated by co-purification with brain microtubules, although mass identification of MAPs had to wait until lab members Edouard Pesquet and Paul Derbyshire used proteomics to identify >600 Arabidopsis MAPs ( Derbyshire et al., 2015 ). Before the advent of proteomics, we isolated MAPs from detergent-extracted carrot ( Daucus carota ) cytoskeletons. In this way, Chan et al. (1999) purified a triplet of ca 65 kDa proteins that bundled brain microtubules in vitro, reproducing the characteristic 30 nm cross-bridges that maintained the parallelism of cortical microtubules in planta. These proved to be a new class of plant proteins, unrelated to classical brain MAPs ( Smertenko et al., 2000 ). Because only one MAP65 isoform was expressed in carrot cells arrested in interphase, we suggested that other family members must attach to other microtubule arrays in dividing cells ( Lloyd and Chan, 2004 ). Mutagenesis of the consensus phosphorylation site of cyclin-dependent kinase on Arabidopsis GFP-AtMAP65-1 (of a nine-gene-member family) showed that labeling is downregulated at metaphase but returns to the central spindle at anaphase ( Mao et al., 2005 ). It would appear that different MAP65 isoforms are expressed at different stages of the cell cycle and that the isoform that cross-links microtubules during interphase is switched off during metaphase by cell cycle-regulated phosphorylation.
To screen for plant-specific MAPs that (1) would escape bioinformatic searches based on motifs from animal MAPs and (2) might not have bound to animal microtubules, we used Taxol to polymerize endogenous tubulin from Arabidopsis suspension cells. Co-purifying proteins were analyzed by mass spectrometry, yielding 55 candidates, amongst which was the five-member MAP70 family, found only in plants ( Korolev et al., 2005 ). Another protein, occurring in plants and trypanosomes, was subsequently characterized by Buschmann et al. (2006) . This 187 kDa protein, AIR9, decorates the PPB but then disappears during metaphase ( Figure 5 ). However, AIR9 reappears as a ring at the predicted cortical division site the instant the phragmoplast makes contact, suggesting that AIR9 associates with a molecular imprint deposited by the PPB.

MAPs during mitosis. When expressed in tobacco BY-2 cells, GFP-AIR9 labels the PPB (A) but disappears from the cortex at metaphase (B). It reappears in a thin line at the cortex (arrowed in C) when the phragmoplast makes contact. In contrast, GFP-KCBP is detected in the PPB (D) but remains at the cortical division site in a particulate form during metaphase (E) and cytokinesis (F). Scale bar = 20 µm. Reproduced from Buschmann et al. (2015) .
In searching for its binding partners, Henrik found that AIR9 interacts with the kinesin-like calmodulin-binding protein KCBP, a microtubule motor protein that stays in the cortical division site throughout mitosis ( Buschmann et al., 2015 ; Figure 5 ). The ability of Arabidopsis KCBP to bind the cortex is conferred by its MyTH4-FERM domain, an evolutionarily conserved module known to link the actin cytoskeleton to microtubules. We hypothesized that minus-end-directed KCBP anchored at the former PPB site could “reel in” the plus ends of microtubules growing out from the phragmoplast. (KCBP’s connection with actin is also interesting, since actin filaments connect the dividing nucleus to the cortex [ Traas et al., 1987 ])
Another positive marker of the cortical division site is TANGLED ( Walker et al., 2007 ) and, in a recent collaboration with Henrik, Carolyn Rasmussen’s lab ( Mir et al., 2018 ) made synthetic air9 tan1 double mutants to establish that both proteins are required for normal division plane alignment. The longstanding suspicion that the PPB leaves behind an imprint—a molecular tidemark—has now been confirmed, and it would appear that AIR9, KCBP, and TAN1 are key parts of that complex.
David Robinson: ER exit sites, ER import sites, and bidirectional transport in the early secretory pathway
David Robinson’s work has focused on vesicle trafficking from the ER to the Golgi apparatus (anterograde pathway) as well as that from the Golgi to the ER (retrograde pathway). His group extended and expanded on studies carried out in yeast, and, in addition to describing key aspects of the plant secretory pathway, he identified several unique aspects. These include the findings that the plant Golgi apparatus is extraordinarily dynamic and that anterograde trafficking may occur at places where the ER and Golgi are in close proximity.
I became fascinated with the plant Golgi apparatus since my days as a postdoc with Peter Ray (Stanford). After an interlude where I devoted my attention to the problem of microtubules and cellulose microfibril orientation, I returned to various aspects of the plant secretory pathway. These led to the first studies on endocytosis and clathrin-coated vesicle (CCV) isolation. The discovery of COP-coated vesicles in yeast and animals prompted us in the 1990s to ascertain their existence in plants. This is now indisputable. In the following section, I give a personalized account of the contribution of my group to the problems surrounding ER–Golgi vesicle-mediated cycling.
Studies on the plant Golgi apparatus were really given a boost when my colleague and friend, the late Chris Hawes, noted that the Golgi stacks of plant cells moved along the surface of tubular strands of ER ( Boevink et al., 1998 ). However, even more interesting were the observations that this movement was intermittent (“stop-and-go”) and dependent on intact actin filaments. This immediately prompted the questions: where on the surface of the ER does ER export take place, and is export a continuous process or restricted to the stationary periods of the Golgi stacks? At that time, studies on animal and yeast cells had already established the existence and anterograde trafficking properties of COPII-coated vesicles, as well retrograde trafficking of COPI-coated vesicles (see Béthune and Wieland, 2018 for references). However, at the same time, antibodies against plant COPI and COPII-homolog proteins became available ( Movafeghi et al., 1999 ), opening the way to the subcellular localization of COPI vesicles in plants ( Pimpl et al., 2000 ).
My group next turned to the challenge of subcellular localization of COPII binding on the ER for anterograde transport, sites then termed ERES (ER exit sites). To this end, two experimental approaches were possible: immunofluorescence and the detection of transiently expressed fluorescently tagged [(X)-FP] COPII proteins. The first procedure applied to suspension-cultured tobacco BY-2 cells produced a uniform, punctate labeling of the ER, with the number of punctae greatly in excess of the number of visualized Golgi stacks ( Yang et al., 2005 ). In contrast, daSilva et al. (2004) working with tobacco epidermal cells demonstrated a high degree of colocalization between the Golgi markers ST-GFP and ERD2-GFP and the GTPase SAR1-YFP (SAR1 is a GTPase that had previously been shown to be required for vesicle transport from the ER). This distribution pattern was later confirmed by examining YFP fusions to other COPII proteins (SEC23 and SEC24; Hanton et al., 2007 ).
The “Secretory Unit Concept” ( daSilva et al., 2004 ) was therefore formulated as a consequence of the colocalization of Golgi and COPII markers. It describes a specific domain of the ER, containing ERES, which together with an overlying Golgi stack, constitutes a mobile secretory aggregate (see “Brandizzi” section in Robinson et al., 2015 , for a discussion; see also Figure 6 ). This is a distinguishing feature of the endomembrane system of plant cells, since the Golgi apparatus in animal cells is usually seen as a fused ribbon of stacks in a perinuclear position that is nonmotile ( Saraste and Prydz, 2019 ).

Cartoon portraying vesicle-mediated anterograde (COPII vesicles) and retrograde (COPI vesicles) between the ER and Golgi. Vesicle trafficking is accomplished by the sequential operation of long-range tethering complexes (in the case of retrograde trafficking to the ER, the complex includes the plant homolog to TIP20 of the yeast Dsl1 COPI-tethering complex, and in the case of anterograde trafficking to the Golgi, the cis-Golgi tethering complex includes the component p115), and short-range SNARE vesicle fusion complexes. In the case of the ER, this is the cis-SNARE vesicle fusion complex (of which SYP-72 is a component). In this model, ERES and ERIS are in close proximity in a domain of the ER whose size approximates the diameter of an overlying Golgi stack. Golgi stacks are in tight association with the ER via a joint scaffolding matrix of tethering factors. It is proposed that as Golgi stacks move, they capture individual COPII vesicles released from ERES, and simultaneously release COPI vesicles. Both types of vesicles accumulate in the restricted ER–Golgi interface. When the Golgi stacks temporarily stop moving (docking phase), fusion of COPI and COPII vesicles to their respective target membranes takes place. Adapted from Lerich et al. (2012) Figure 13A.
There are still many unknowns concerning the function of the secretory unit. For example, there is an ongoing discussion as to whether the mediators of bidirectional protein traffic between the ER and cis-Golgi are bona fide vesicles (COPI and COPII), or whether transport is accomplished through direct tubular connections that are partially coated with COPI or COPII proteins (see “Hawes” section in Robinson et al., 2015 ; Brandizzi, 2018 : Robinson, 2020 ). It also remains unclear whether bidirectional transport is a continuous process or restricted to stationary Golgi stacks. What is clear, through the use of optical tweezers, is that the secretory unit is an extremely stable complex. This stability is not indicative of direct connections between the ER and Golgi membranes, but rather is due to a tethering complex based on the golgin AtCASP ( Osterrieder et al., 2017 ).
COPI-mediated retrograde trafficking has been well-studied in animal cells and involves an interaction of γ-COP with the cytoplasmic tails of p24 dimers ( Béthune and Wieland, 2018 ). There are also interactions between α- and β′-COP and dilysine motifs on retrograde cargo molecules such as the KDEL receptor. Plants also possess p24 proteins, and it is generally accepted that KDEL-receptor–ligand complexes destined for retrieval to the ER interact with both the COPI GTPASE ARF1 and p24 proteins ( Pastor-Cantizano et al., 2016 ). The receptor for KDEL-ligands is known as ERD2 in yeast and plants and has a putative COPI dilysine cluster. It is thought that ERD2 cycles between the ER and the cis-Golgi, releasing ligands into the lumen of the ER after COPI-vesicle fusion due to the neutral pH in this compartment. After having returned to the cis-Golgi via COPII-vesicles, the low pH in the cis-cisternae causes ERD2 to bind KDEL ligands and also leads to a conformational change in the ERD2 receptor that exposes lysine residues, thereby creating a COPI-binding motif ( Robinson and Aniento, 2020 ).
Although it cycles between the cis-Golgi and the ER, ERD2 curiously locates predominantly to the cis-Golgi, being detected in the lumen of the ER only after over-expression of KDEL-ligands. In a model proposed by Robinson and Aniento (2020) , it is assumed that under normal conditions, ERD2 is rapidly exported out of the ER. It is therefore only transitorily present in the ER and is restricted to a disc-like domain immediately underneath the first cis-Golgi cisterna. Its presence there makes it difficult to distinguish from the cis-Golgi population of ERD2 in face views of the Golgi under a confocal laser-scanning microscope.
The scenario just described implies that ER import sites are closely associated with ERES and lie immediately beneath the cis-Golgi. That this is indeed so comes from studies to pinpoint ER-located tethering factors and SNAREs responsible for COPI-vesicle tethering and subsequent fusion, respectively ( Lerich et al., 2012 ). An (X)FP-tagged plant homolog to TIP20 of the yeast Dsl1 COPI-tethering complex perfectly colocalizes with Golgi stacks irrespective of their motile status. However, in contrast, the Qc-SNARE SYP72, whose overexpression has no effect on ER–Golgi trafficking, only colocalizes with Golgi markers when the stacks are immobilized through actin depolymerization. These findings suggest that punctate SYP72-YFP signals on the ER, which do not colocalize with Golgi stack markers, represent predetermined docking sites, and that COPI-vesicle fusion with the ER is restricted to periods when Golgi stacks are stationary (see Figure 6 ; for details of the actual labeling, the reader is referred to Lerich et al., 2012 ). The most gratifying aspect of this publication is that the same strategy was employed most recently by to define “bidirectional transport portals” on the ER of the yeast Pichia pastoris : a rare case where a plant publication has provided a template for animal/yeast studies on vesicle-mediated protein transport.
Andrew Staehelin: Elucidation of the nanoscale architecture of cells—highlights of a long journey
Andrew Staehelin developed cutting-edge cryofixation methods that he used to characterize novel features of plant cell structures through electron microscopy. His work addresses cell plate formation during cytokinesis, thylakoid structure, as well as various other topics. Here he highlights early work on cellulose-synthesizing rosette complexes, and structure/function studies of the plant Golgi apparatus.
The long-term goal of my career was to produce high-resolution 3D models of cellular organelles and cytoskeletal systems in their natural state. In turn, I hoped that this information would provide a structural framework for understanding molecular, biochemical, and fluorescent microscope data produced by other researchers.
As a graduate student in the 1960s, I recognized that the principal limitation of biological electron microscopy was not the resolution limit of the electron microscopes but the inability of researchers to preserve the native architecture of cells for viewing in an electron microscope. In particular, I recognized the limitations of chemical fixatives and that cryofixation provided the only alternative fixation method.
The development of propane jet-freezing of turgid cells led to the discovery of novel membrane structures
The development of a propane jet-freezing apparatus capable of vitrifying up to 40-µm thick samples (described in Gilkey and Staehelin, 1986 ) yielded our first images of freeze-fractured vitrified cells. This enabled us to address the question of how cellulose fibrils are produced by plants and green algae capable of producing primary and secondary cell walls. Using growing semi-cells of the unicellular green alga Micrasterias as an experimental system, we demonstrated that cellulose fibrils were synthesized by plasma membrane rosette particle complexes ( Giddings et al., 1980 ). Single rosettes were shown to produce ∼5-nm fibrils in primary walls, whereas rows of rosettes in arrays gave rise to larger diameter fibrils in secondary walls. These complexes were delivered in a preassembled form to the plasma membrane. More primitive algae possess linear particle complexes similar to those found in the bacterium Acetobacter xylinum ( Brown and Montezinos,1976 ).
How does turgor pressure affect vesicle-mediated secretion and membrane recycling? Using sycamore, maple, and carrot suspension-cultured cells as experimental systems, we observed that during exocytosis, the vesicles collapsed into disc-shaped membrane appendages because their membranes were unable to be absorbed into the turgid plasma membrane ( Staehelin and Chapman, 1987 ). Recycling of the appendage membranes involves the formation of horseshoe-shaped membrane infoldings that become cupped by closely associated ER membranes until the excess membrane has been resorbed, presumably by a lipid-hopping mechanism ( Lev, 2010 ). The proteins were retrieved by CCVs.
High-pressure freezing enabled us to develop functional, nanoscale models of ER, Golgi, and TGN cisternae
Thin section studies of high-pressure frozen Arabidopsis and tobacco root tip cells demonstrated that the Golgi stacks consisted of three types of structurally distinguishable cisternae, cis - , medial-, and trans-Golgi cisternae ( Staehelin et al., 1990 ). These structural differences suggested that they were functionally different. We tested this hypothesis using antibodies against sugar epitopes of secreted polysaccharides and glycoproteins in cryofixed Golgi. The results confirmed our hypothesis. For example, antibodies against the backbone and the sidechains of xyloglucans bound only to trans-Golgi and TGN cisternae ( Zhang and Staehelin, 1992 ), whereas with anti-native α-1,2 mannosidase antibodies, 90% of the label was in medial cisternae ( Donohoe et al., 2013 ).
Studies of the trafficking of fluorescently labeled Golgi stacks in living tobacco BY-2 cells led to the formulation of the “stop-and-go” model of Golgi stack trafficking along ER-associated actin filaments ( Nebenführ et al., 1999 ). What causes moving Golgi stacks to stop? Where do they stop? The discovery of a ribosome-excluding and Golgi-TGN encompassing scaffold/matrix ( Figure 7A ) provided the first clue. This scaffold was shown to originate as an external layer on budding COPII vesicles at ER exit sites and to contain the Atp115 scaffolding protein ( Kang and Staehelin, 2008 ). Once assembled, this scaffold enabled budding COPII vesicles to capture passing Golgi by attaching to the cis side of the Golgi scaffold. After binding, the docked, wiggling Golgi pluck the COPII vesicles from the ER, which are then incorporated into growing cis cisternae. Simultaneously, the COPII scaffold molecules become integrated into the Golgi scaffold. These results led to the formulation of the revised “stop-pluck-and-go” model of Golgi trafficking ( Staehelin and Kang, 2008 ).

3D tomographic models of a dividing Arabidopsis Golgi stack and TGN cisternae. A, Interactions between the COPII scaffolds (arrowheads) and the cis - side of the Golgi/TGN matrix/scaffold connect ER and Golgi. The dividing Golgi possesses two sets of cis- and medial-cisternae (green, purple, and yellow) held together by the undivided trans-Golgi cisterna (pink). B, Face-on views of the cis - cisternae of the dividing Golgi shown in (A). The C1 and C1′ (orange) and the C2 and C2′ cisternae (green) are cisternal assembly intermediates. COPII vesicles are seen around the margins of these cisternae. C, 3D model images of a Golgi stack with a Golgi-associated TGN (TGN1) and a free, fragmenting TGN (TGN2). C and D, Release of the secretory and CCVs and residual cisternal fragments (asterisk, red) occurs simultaneously.
Prior to the introduction of cryofixation to plant cell biology, electron microscopists searched in vain for COPII vesicles in chemically fixed cells. After visualizing COPII vesicles in high-pressure frozen root tip cells ( Craig and Staehelin, 1988 ), we focused our efforts on characterizing the distribution of COPII and retrograde COPI-type Golgi vesicles around Golgi stacks. This work led to the discovery of two subtypes of COPI vesicles, COPIa and COPIb ( Donohoe et al., 2007 ). COPIa and COPII vesicles were seen exclusively between ER exit sites and the cis-cisternae of Golgi stacks, whereas the COPIb vesicles were observed only around the medial- and trans-Golgi and TGN cisternae. This suggested that COPIa vesicles were involved in membrane recycling between the cis-cisternae and the ER, and the COPIb vesicles in recycling between medial-, trans-Golgi, and TGN cisternae ( Donohoe et al., 2007 ). Quantitative analysis of cisternal membrane recycling demonstrated furthermore that during the transformation of the trans-most Golgi cisterna to a TGN cisterna, between 30% and 35% of the membrane is recycled ( Kang et al., 2011 ).
Two central predictions of the cisternal progression model of Golgi trafficking are that new Golgi cisternae are assembled on the cis-side of the stacks, while old cisternae are shed on the trans-side ( Day et al., 2013 ). Electron tomography enabled us to follow cisternal assembly from the initial fusion of two COPII vesicles on the surface of the existing cis-most cisterna followed by the growth and maturation of a new cis-cisterna ( Figure 7B; Donohoe et al., 2013 ). In turn, the trans-most Golgi cisternae were observed to peel off the stacks, mature into TGN cisternae, and release the secretory and CCVs by cisternal fragmentation ( Figure 7, C and D ; Kang et al., 2011 ).
Further analysis of the de novo assembly of the cis-Golgi cisternae demonstrated that cis-cisternae are strictly cisternal assembly compartments from which escaped ER proteins are returned to the ER via COPIa vesicles. Activation of the enzymatic activities occurs only in the medial cisternae ( Donohoe et al., 2013 ).
Looking back, I never imagined that the decision to focus my career on the development of improved cryofixation and specimen preparation methods would lead to such a wealth of novel insights into plant cell structures and functions. During this pursuit, I was astonished that it took 15 years for the electron microscopy research community to accept the high-pressure freezing technique. Now cryofixation is considered an essential tool for producing nanoscale 3D reconstructions of cellular membrane and cytoskeletal systems by means of electron tomography.
Concluding remarks
As illustrated by the retrospectives in this review, the plant cell biology field has seen tremendous progress in the past decades, truly a bountiful harvest. In part, investigations are inspired by the unique nature of walled plant cells compared to other multicellular eukaryotes. In those cases, we have learned how plants have solved problems toward developing multicellular bodies differently from animals of fungi. In other cases though, plant cells have proven to be superior model systems for general eukaryotic cellular processes. The “vignettes” in this review show how deep investigation, along with implementing or developing new experimental approaches, can illuminate fundamental properties of plant cells, with impacts reaching to all fields of plant science. We hope these stories will continue to inspire future investigations and breakthroughs in the exciting field of plant cell biology, where so much is yet to be learned.
Acknowledgments
Clive Lloyd acknowledges the key contributions of lab members and collaborators to the work reported here. Henrik Buschmann is thanked for supplying Figure 5 and for discussions.
Work in Alice Cheung’s lab over the years has been supported by the US National Science Foundation (NSF), US Department of Agriculture (USDA), National Institutes of Health (NIH), and Department of Energy (DOE). Work on expansins in Daniel Cosgrove.’s lab was supported by the US Department of Energy (grant no. DE-FG2-84ER13179), and work on cell wall structure was supported as part of the Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences (award no. DE-SC0001090). Research in Ikuko Hara-Nishimura’s lab is supported by Grant-in-Aid for Scientific Research (no. 15H05776) and Specially Promoted Research of Grant-in-Aid for Scientific Research (no. 22000014) from the Japan Society for the Promotion of Science and by the Hirao Taro Foundation of KONAN GAKUEN for Academic Research. Research on cytokinesis in Gerd Jürgens’ lab has been funded by the Deutsche Forschungsgemeinschaft. Much of Clive Lloyd’s work was supported by the Biotechnology and Biosciences Research Council, UK. Work on plant cell biology in Dolf Weijers’ lab is funded by the European Research Council (ERC) through an Advanced Grant (DIRNDL—contract number 833867).
Conflict of interest statement . The authors do not have any competing interests to disclose.
Contributor Information
Alice Y Cheung, Department of Biochemistry and Molecular Biology, Molecular Cell Biology Program, Plant Biology Program, University of Massachusetts, Amherst, Massachusetts 01003, USA.
Daniel J Cosgrove, Department of Biology, Penn State University, University Park, Pennsylvania 16802, USA.
Ikuko Hara-Nishimura, Faculty of Science and Engineering, Konan University, Kobe 658-8501, Japan.
Gerd Jürgens, ZMBP-Developmental Genetics, University of Tuebingen, Tuebingen 72076, Germany.
Clive Lloyd, Department of Cell and Developmental Biology, John Innes Centre, Norwich NR4 7UH, UK.
David G Robinson, Centre for Organismal Studies, University of Heidelberg, Heidelberg D-69120, Germany.
L Andrew Staehelin, Department of Molecular, Cellular and Developmental Biology, University of Colorado, Boulder, Colorado 80309-0347, USA.
Dolf Weijers, Laboratory of Biochemistry, Wageningen University, Wageningen 6708WE, the Netherlands.
All contributing authors conceived and wrote their individual part, as marked in each section, prepared figures and commented on a complete draft, and prepared and compiled by D.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors ( https://academic.oup.com/plcell ) is: Dolf Weijers ( [email protected] ).
- Assaad FF, Huet Y, Mayer U, Jürgens G (2001) The cytokinesis gene KEULE encodes a Sec1 protein which binds the syntaxin KNOLLE . J Cell Biol 152 : 531–544 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM (2015) A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly . Science 349 : 1111–1114 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Béthune J, Wieland FT (2018) Assembly of COPI and COPII vesicular coat proteins on membranes . Annu Rev Biophys 47 : 63–83 [ PubMed ] [ Google Scholar ]
- Boevink P, Oparka K, Cruz SS, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on anactin/ER network . Plant J 15 : 441–447 [ PubMed ] [ Google Scholar ]
- Brandizzi F (2018) Transport from the endoplasmic reticulum to the Golgi in plants: where are we now? Semin Cell Dev Biol 80 : 94–105 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Brown RM Jr, Montezinos D (1976) Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane . Proc Natl Acad Sci USA 73 :143–147 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW (2006) Microtubule-associated AIR9 recognises the cortical division site at preprophase and cell-plate insertion . Curr Biol 16 : 1938–1943 [ PubMed ] [ Google Scholar ]
- Buschmann H, Dols J, Kopischke S, Peña EJ, Andrade-Navarro MA, Heinlein M, Szymanski DB, Zachgo S, Doonan JH, Lloyd CW (2015) Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain . J Cell Sci 128 : 2033–2046 [ PubMed ] [ Google Scholar ]
- Chan J, Crowell E, Eder M, Calder G, Bunnewell S, Findlay K, Vernhettes S, Höfte H, Lloyd CW (2010) The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls . J Cell Sci 123 : 3490–3495 [ PubMed ] [ Google Scholar ]
- Chan J, Calder GM, Fox S, Lloyd CW (2007) Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells . Nat Cell Biol 9 : 171–175 [ PubMed ] [ Google Scholar ]
- Chan J, Calder GM, Doonan JH, Lloyd CW (2003) EB1 reveals mobile microtubule nucleation sites in Arabidopsis . Nat Cell Biol 5 : 967–971 [ PubMed ] [ Google Scholar ]
- Chan J, Jensen CG, Jensen LCW, Bush M, Lloyd CW (1999) The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules . Proc Natl Acad Sci USA 96 : 14931–14936 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chen C, Wong E, Vidali L, Estavillo A, Hepler PK, Wu HM, Cheung AY (2002) The regulation of actin organization by actin depolymerizing factor (ADF) in elongating pollen tubes . Plant Cell 14 : 2175–2190 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chen CY, Cheung AY, Wu HM (2003) Actin depolymerizing factor mediates Rac/Rop GTPase regulated pollen tube growth . Plant Cell 15 : 237–249 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chen CY, Zheng WG, Cheung AY, Wu HM (2013) Pollen germination activates the apical membrane located RAC/ROP GTPase switch . Mol Plant 6 : 1358–1361 [ PubMed ] [ Google Scholar ]
- Cheung AY, Chen C, Glaven R, Vidali L, Hepler PK, Wu HM (2002) Rab2 regulate vesicular transport between endoplasmic reticulum and Golgi bodies and is important for pollen tube elongation . Plant Cell 14 : 945–962 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cheung AY, Niroomand S, Zou Y, Wu HM (2010) A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes . Proc Natl Acad Sci USA 107 : 16390–16395 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cheung AY, Wang H, Wu HM (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth . Cell 82 : 383–393 [ PubMed ] [ Google Scholar ]
- Cheung AY, Wu HM (2004) Over-expression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane . Plant Cell 16 : 257–269 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cheung AY, Wu HM (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes . Annu Rev Plant Biol 59 : 547–572 [ PubMed ] [ Google Scholar ]
- Cheung AY, Wu HM (2011) THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr Opin Plant Biol 14 : 632–641 [ PubMed ] [ Google Scholar ]
- Cleland R, Haughton PM (1971) The effect of auxin on stress relaxation in isolated Avena coleoptiles . Plant Physiol 47 : 812–815 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cosgrove DJ (1985) Cell wall yield properties of growing tissues. Evaluation by in vivo stress relaxation . Plant Physiol 78 : 347–356 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cosgrove DJ (1987b) Wall relaxation in growing stems: comparison of four species and assessment of measurement techniques . Planta 171 : 266–278 [ PubMed ] [ Google Scholar ]
- Cosgrove DJ (1989) Characterization of long-term extension of isolated cell-walls from growing cucumber hypocotyls . Planta 177 : 121–130 [ PubMed ] [ Google Scholar ]
- Cosgrove DJ (1993) Water uptake by growing cells: an assessment of the controlling roles of wall relaxation, solute uptake, and hydraulic conductance . Int J Plant Sci 154 : 10–21 [ PubMed ] [ Google Scholar ]
- Cosgrove DJ (2020) Non-enzymatic action of expansins . J Biol Chem 295 : 6782. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Craig S, Staehelin LA (1988) High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems . Eur J Cell Biol 146 : 80–93 [ PubMed ] [ Google Scholar ]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F (2004) Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells . Plant Cell 16 : 1753–1771. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Day KJ, Staehelin LA, Glick BS (2013) A three-stage model of Golgi structure and function . Histochem Cell Biol 140 : 239–249 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- de Graaf BHJ, Cheung AY, Andreyeva T, Levasseur K, Kieliszewski M, Wu HM (2005) Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused Pollen tube growth in tobacco . Plant Cell 17 : 2564–2579 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Derbyshire P, Ménard D, Green P, Saalbach G, Buschmann H, Lloyd CW, Pesquet E (2015) Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in arabidopsis . Plant Cell 27 : 2709–2726 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Doblas VG, Gonneau M, Hofte H (2018) Cell wall integrity signaling in plants: malectin domain kinases and lessons from other kingdoms . Cell Surf 3 : 1–11 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Donohoe BS, Kang BH, Staehelin LA (2007) Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi . Proc Natl Acad Sci USA 104 : 163–168 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Donohoe BS, Kang BH, Gerl M, Gergeley ZR, McMichael CM, Bednarek S, Staehelin LA (2013) Cis -Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae . Traffic 14 : 551–567 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Du M, Spalding EP, Gray WM (2020) Rapid auxin-mediated cell expansion . Ann Rev Plant Biol . 71 : 379–402 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development . Proc Natl Acad Sci USA 107 : 17821–17826 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY (2014) Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis . Nat Commun 5 : 3129.21 [ PubMed ] [ Google Scholar ]
- Duan Q, Liu MCJ, Kita D, Jordan S, Yeh FLJ, Yvon R, Carpenter H, Federico AN, Garcia-Valencia L, Eyles SJ, et al. (2020) FERONA controls pectin- and nitric oxide-mediated male-female interaction . Nature 579 : 561–566 [ PubMed ] [ Google Scholar ]
- El Kasmi F, Krause C, Hiller U, Stierhof YD, Mayer U, Conner L, Kong L, Reichardt I, Sanderfoot AA, Jürgens G (2013) SNARE complexes of different composition jointly mediate membrane fusion during cytokinesis in Arabidopsis . Mol Biol Cell 24 : 1593–1601 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Enami K, Ichikawa M, Uemura T, Kutsuna N, Hasezawa S, Nakagawa T, Nakano A, Sato MH (2009) Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana . Plant Cell Physiol . 50 : 280–289 [ PubMed ] [ Google Scholar ]
- Escobar-Restrepo, J-M, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang W-C, Grossniklaus U (2007) The FERONIA receptor kinase mediates male-female interactions during pollen tube reception. Science 317 : 656–660 [ PubMed ] [ Google Scholar ]
- Feng W, Kita D, Peaucelle A, Doan V, Duan Q, Liu MC, Maman J, Yvon R, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell wall integrity during salt stress through Ca 2+ signaling . Curr Biol 28 : 666–675 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ferro-Novick S, Jahn R (1994) Vesicle fusion from yeast to man . Nature 370 , 191–193 [ PubMed ] [ Google Scholar ]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling . Science 358 : 1596–1600 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ge Z, Zhao Y, Liu MC, Wang L, Zhong S, Hou S, Jian J, Liu T, Huang Q, Xiao J, et al. (2019) LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity . Curr Biol 29 : 3256–3265 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Giddings TH, Brower DL, Staehelin LA (1980) Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls . J Cell Biol 84 : 327–339 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Gilkey JC, Staehelin LA (1986) Advances in ultrarapid freezing for the preservation of cellular ultrastructure . J El Microsc Tech 3 :177–212 [ Google Scholar ]
- Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F (2007) De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated . Plant Physiol 143 : 1640–1650 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hara-Nishimura I, Nishimura M, Akazawa T (1985) Biosynthesis and intracellular transport of 11S globulin in developing pumpkin cotyledons . Plant Physiol 77 : 747–752 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hara-Nishimura I, Takeuchi Y, Nishimura M (1993) Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni . Plant Cell 5 : 1651–1659 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion . Science 343 : 408–411 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death . Science 305 : 855–858 [ PubMed ] [ Google Scholar ]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I (2009) A novel membrane-fusion-mediated plant immunity against bacterial pathogens . Gene Dev 23 : 2496–2506 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G (2001) Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis . J Cell Biol 155 : 239–250 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hematy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Baizergue S, Pelletier S, Renou JP, Hofte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis . Curr Biol 17 : 922–993 [ PubMed ] [ Google Scholar ]
- Hepler PK, Newcomb EH (1967) Fine structure of cell plate formation in the apical meristem of Phaseolus roots . J Ultrastruc Res 19 : 53–71. [ PubMed ] [ Google Scholar ]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation . Nat Commun 5 : 3978. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hush JM, Wadsworth P, Callaham DA, Hepler PK (1994) Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching . J Cell Sci 107 : 775–784 [ PubMed ] [ Google Scholar ]
- Jahn R, Scheller RH (2006) SNAREs – engines for membrane fusion . Nat Rev Mol Cell Biol 7 : 631–643 [ PubMed ] [ Google Scholar ]
- Jones AM, Chory J, Dangl JL, Estelle M, Jacobsen SE, Meyerowitz EM, Nordborg M, Weigel D (2008) The impact of Arabidopsis on human health: diversifying our portfolio . Cell 133 : 939–943 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Kang BH, Staehelin LA (2008) ER-to-Golgi transport by COPII vesicles in Arabidopsis thaliana involves a ribosome-excluding scaffold that is transferred with the vesicles to the Golgi matrix . Protoplasma 234 : 51–64 [ PubMed ] [ Google Scholar ]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b and PI-4Kβ1 labeled trans - Golgi network compartments in Arabidopsis . Traffic 12 : 313–329 [ PubMed ] [ Google Scholar ]
- Karnahl M, Park M, Mayer U, Hiller U, Jürgens G (2017) ER assembly of SNARE complexes mediating formation of partitioning membrane in Arabidopsis cytokinesis . eLife 6 : e25327. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Karnahl M, Park M, Krause C, Hiller U, Mayer U, Stierhof YD, Jürgens G (2018) Functional diversification of Arabidopsis SEC1-related SM proteins in cytokinetic and secretory membrane fusion . Proc Natl Acad Sci USA 115 : 6309–6314 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Kerff F, Amoros A, Herman R, Sauvage E, Petrella S, Filee P, Charlier P, Joris B, Tabuchi A, Nikolaidis N, et al. (2008) Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization . Proc Natl Acad Sci USA 105 : 16876–16881 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Korolev AV, Chan J, Naldrett MJ, Doonan JH, Lloyd CW (2005) Identification of a novel family of 70kDa microtubule-associated proteins in Arabidopsis cells . Plant J 42 : 547–555 [ PubMed ] [ Google Scholar ]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin . J Cell Biol 139 : 1485–1493 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Lerich A, Hillmer S, Langhans M, Scheuring D, van Bentum P, Robinson DG (2012) ER import sites and their relationship to ER exit sites: a new model for bidirectional ER-golgi transport in higher plants . Front Plant Sci 3 : 143. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Lev S (2010) Non-vesicular lipid transport by lipid-transfer proteins and beyond . Nat Rev Mol Cell Biol 11 : 739–750 [ PubMed ] [ Google Scholar ]
- Li C, Wu HM, Cheung AY (2016) FERONIA and her pals: functions and mechanisms . Plant Physiol 171 : 2379–2391 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Li C, Yeh F, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis . eLife 4 : e06587 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Liu C, Shen L, Xiao Y, Vyshedsky D, Peng C, Sun X, Liu Z, Cheng L, Zhang H, Han Z, et al. (2021) Pollen PCP-B peptides unlock a stigma peptide-receptor kinase gating mechanism for pollination . Science 372 : 171–175 [ PubMed ] [ Google Scholar ]
- Lloyd C, Chan J (2004) Microtubules and the shape of plants to come . Nat Rev Mol Cell Biol 5 : 13–22 [ PubMed ] [ Google Scholar ]
- Lloyd CW, Slabas AR, Powell AJ, Macdonald G, Badley RA (1979) Cytoplasmic microtubules of higher plant cells visualised with anti-tubulin antibodies . Nature 279 : 239–241 [ Google Scholar ]
- Lukowitz W, Mayer U, Jürgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product . Cell 84 : 61–71 [ PubMed ] [ Google Scholar ]
- Mao G, Chan J, Calder G, Doonan JH, Lloyd CW (2005) Modulated targeting of GFP-AtMAP65-1 to central spindle microtubules during division . Plant J 43 : 469–478 [ PubMed ] [ Google Scholar ]
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G (1991) Mutations affecting body organization in the Arabidopsis embryo . Nature 353 : 402–407 [ Google Scholar ]
- McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants . Plant Cell 4 : 1425–1433 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- McQueen-Mason SJ, Cosgrove DJ (1995) Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding . Plant Physiol 107 : 87–100 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mir R, Morris VH, Buschmann H, Rasmussen CG (2018) Division plane orientation defects revealed by a synthetic double mutant phenotype . Plant Physiol 176 : 418–431 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth . Nature 312 : 237–242 [ PubMed ] [ Google Scholar ]
- Movafeghi A, Happel N, Pimpl P, Tai GH, Robinson DG (1999) Arabidopsis Sec21p and Sec23p homologs. Probable coat proteins of plant COP-coated vesicles . Plant Physiol 119 : 1437–1446 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Narvaez-Barragan DA, Tovar-Herrera OE, Segovia L, Serrano M, Martinez-Anaya C (2020) Expansin-related proteins: biology, microbe-plant interactions and associated plant-defense responses . Microbiology (Reading) 166 : 1007–1018 [ PubMed ] [ Google Scholar ]
- Nebenführ A, Gallagher L, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system . Plant Physiol 121 : 1127–1141 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Nikolaidis N, Doran N, Cosgrove DJ (2014) Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion . Mol Biol Evol 31 : 376–386 [ PubMed ] [ Google Scholar ]
- Okamoto K, Ueda H, Shimada T, Tamura K, Kato T, Tasaka M, Morita MT, Hara-Nishimura I (2015) Regulation of organ straightening and plant posture by an actin–myosin XI cytoskeleton . Nat Plants 1 : 15031 DOI: 10.1038/NPLANTS.2015.1031. [ PubMed ] [ Google Scholar ]
- Okamoto T, Shimada T, Hara-Nishimura I, Nishimura M, Minamikawa T (2003) C-terminal KDEL sequence of a KDEL-tailed cysteine proteinase (sulfhydryl-endopeptidase) is involved in formation of KDEL vesicle and in efficient vacuolar transport of sulfhydryl-endopeptidase . Plant Physiol 132 : 1892–1900 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Osterrieder A, Sparkes IA, Botchway SW, Ward A, Ketelaar T, deRuijter N, Hawes C (2017) Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-Golgi apparatus tethering . J Exp Bot 68 : 3339–3350 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pastor-Cantizano N, Montesinos JC, Bernat-Silvestre C, Marcote MJ, Aniento F (2016) p24 family proteins: key players in the regulation of trafficking along the secretory pathway . Protoplasma 253 : 967–985 [ PubMed ] [ Google Scholar ]
- Paradez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules . Science 312 : 1491–1495 [ PubMed ] [ Google Scholar ]
- Park M, Touihri S, Müller I, Mayer U, Jürgens G (2012) Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis . Dev Cell 22 : 989–1000 [ PubMed ] [ Google Scholar ]
- Park M, Krause C, Karnahl M, Reichardt I, El Kasmi F, Mayer U, Stierhof YD, Hiller U, Strompen G, Bayer M, et al. (2018) Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis . Dev Cell 44 : 500–511 [ PubMed ] [ Google Scholar ]
- Park YB, Cosgrove DJ (2012) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases . Plant Physiol 158 : 1933–1943 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG (2000) In situ localization and in vitro induction of plant COPI coated vesicles . Plant Cell 12 : 2219–2236 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ray PM, Green PB, Cleland R (1972) Role of turgor in plant cell growth . Nature 239 : 163–164 [ Google Scholar ]
- Rayle DL, Cleland RE (1992) The acid growth theory of auxin-induced cell elongation is alive and well . Plant Physiol 99 : 1271–1274 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Reichardt I, Stierhof YD, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G (2007) Plant cytokinesis requires de novo secretory trafficking but not endocytosis . Curr Biol 17 : 2047–2053 [ PubMed ] [ Google Scholar ]
- Reichardt I, Slane D, El Kasmi F, Knöll C, Fuchs R, Mayer U, Lipka V, Jürgens G (2011) Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis . Traffic 12 : 1269–1280 [ PubMed ] [ Google Scholar ]
- Richter S, Kientz M, Brumm S, Nielsen ME, Park M, Gavidia R, Krause C, Voss U, Beckmann H, Mayer U, et al. (2014) Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion . eLife 3 : e02131. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Robinson DG (2020) Plant Golgi ultrastructure . J Microsc 280 : 111–121 [ PubMed ] [ Google Scholar ]
- Robinson DG, Aniento F (2020) A model for ERD2 function in higher plants . Front Plant Sci 11 : 343. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Robinson DG, Brandizzi F, Hawes C, Nakano A (2015) Vesicles versus tubes: is endoplasmic reticulum-golgi transport in plants fundamentally different from other eukaryotes? Plant Physiol 168 : 393–406 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sambade A, Pratap A, Buschmann H, Morris R, Lloyd C (2012) The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl . Plant Cell 24 : 192–201 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily . Genome Biol 6 : 242. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Sampedro J, Guttman M, Li LC, Cosgrove DJ (2015) Evolutionary divergence of beta-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits . Plant J 81 : 108–120 [ PubMed ] [ Google Scholar ]
- Saraste J, Prydz K (2019) A new look at the functional organization of the Golgi ribbon . Front Cell Devel Biol 7 : 1–21 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Schmid M, Simpson D, Kalousek F, Gietl C (1998) A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment . Planta 206 : 466–475 [ PubMed ] [ Google Scholar ]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, Mcqueen-Mason SJ, Shieh M, Cosgrove DJ (1995) Molecular cloning and sequence analysis of expansins - a highly conserved, multigene family of proteins that mediate cell wall extension in plants . Proc Natl Acad Sci USA 92 : 9245–9249 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Shimada T, Takagi J, Ichino T, Shirakawa M, Hara-Nishimura I (2018a) Plant vacuoles . Annu Rev Plant Biol 69 : 123–145 [ PubMed ] [ Google Scholar ]
- Shimada T, Watanabe E, Tamura K, Hayashi Y, Nishimura M, Hara-Nishimura I (2002) A vacuolar sorting receptor PV72 on the membrane of vesicles that accumulate precursors of seed storage proteins (PAC vesicles) . Plant Cell Physiol 43 : 1086–1095 [ PubMed ] [ Google Scholar ]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana . Proc Natl Acad Sci USA 100 : 16095–16100 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Shimada TL, Hayashi M, Hara-Nishimura I (2018b) Membrane dynamics and multiple functions of oil bodies in seeds and leaves . Plant Physiol 176 : 199–207 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Shimada TL, Shimada T, Okazaki Y, Higashi Y, Saito K, Kuwata K, Oyama K, Kato MHU, Nakano A, Ueda T, et al. (2019) HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis . Nat Plants 5 : 1154–1166 [ PubMed ] [ Google Scholar ]
- Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, Asano M, Nishimura M, Hara-Nishimura I (2003) Biosynthetic processing of cathepsins and lysosomal degradation are abolished in asparaginyl endopeptidase-deficient mice . J Biol Chem 278 : 33194–33199 [ PubMed ] [ Google Scholar ]
- Smertenko A, Saleh N, Igarishi H, Mori H, Hauser-Hahn I, Jiang CJ, Sonobe S, Lloyd CW, Hussey PJ (2000) A new class of microtubule-associated proteins in plants . Nat Cell Biol 2 : 750–753 [ PubMed ] [ Google Scholar ]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE (1993) A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion . Cell 75 : 409–418 [ PubMed ] [ Google Scholar ]
- Staehelin LA, Chapman RL (1987) Secretion and membrane recycling in plant cells: novel intermediary structures visualized in ultrarapidly frozen sycamore and carrot suspension culture cells . Planta 171 : 43–57 [ PubMed ] [ Google Scholar ]
- Staehelin LA, Giddings TH Jr, Kiss JZ, Sack FD (1990) Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples . Protoplasma 157 : 75–91 [ PubMed ] [ Google Scholar ]
- Staehelin LA, Kang BH (2008) Nanoscale architecture of ER export sites and of Golgi membranes as determined by electron tomography . Plant Physiol 147 : 1454–1468 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Stegmann M, Monaghan J, Smakowsko-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling . Science 355 : 287–289 [ PubMed ] [ Google Scholar ]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) STOMAGEN positively regulates stomal density in Arabidopsis . Nature 463 : 241–244 [ PubMed ] [ Google Scholar ]
- Tabuchi A, Li LC, Cosgrove DJ (2011) Matrix solubilization and cell wall weakening by beta-expansin (group-1 allergen) from maize pollen . Plant J 68 : 546–559 [ PubMed ] [ Google Scholar ]
- Takagi J, Kimori Y, Shimada T, Hara-Nishimura I (2020) Dynamic capture and release of endoplasmic reticulum exit sites by Golgi stacks in Arabidopsis . iScience 23 : 101265 doi: 10.1016. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis . Curr Biol 23 : 1776–1781 [ PubMed ] [ Google Scholar ]
- Tao LZ, Cheung AY, Wu HM (2002) Plant Rac-like GTPases are activated by auxin and mediate auxin responsive gene expression . Plant Cell 14 : 2745–2760 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Tao LZ, Cheung AY, Nibau SC, Wu HM (2005) RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins . Plant Cell 17 : 369–2383 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Traas JA, Doonan JH, Rawlins DJ, Shaw PJ, Watts PJ, Lloyd CW (1987) An actin network is present in the cytoplasm throughout the cell cycle of carrot cells and associates with the dividing nucleus . J Cell Biol 105 : 387–395 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I (2010) Myosin-dependent ER motility and F-actin organization in plant cells . Proc Natl Acad Sci USA 107 : 6894–6899 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Valdivia ER, Stephenson AG, Durachko DM, Cosgrove D (2009) Class B beta-expansins are needed for pollen separation and stigma penetration . Sexual Plant Reprod 22 : 141–152 [ PubMed ] [ Google Scholar ]
- Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jürgens G, Mayer U (2000) The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis . Curr Biol 10 : 1371–1374 [ PubMed ] [ Google Scholar ]
- Walker KL, Müller S, Moss D, Ehrhardt DW, Smith LG (2007) Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis . Curr Biol 17 : 1827–1836 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Wang H, Wu HM, Cheung AY (1993) Development and pollination regulated accumulation and glycosylation of a stylar transmitting tissue-specific proline rich protein . Plant Cell 5 : 1639–1650 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Wang T, Chen Y, Tabuchi A, Cosgrove DJ, Hong M (2016) The target of beta-expansin EXPB1 in maize cell walls from binding and solid-state NMR studies . Plant Physiol 172 : 2107–2119 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Wu HM, Wang H, Cheung AY (1995) A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower . Cell 82 : 394–403 [ PubMed ] [ Google Scholar ]
- Wu HM, Hazak O, Cheung AY, Yalovsky S (2011) RAC/ROP GTPases and auxin signaling: a perspective . Plant Cell 23 : 1208–1218 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yamada K, Basak AK, Goto-Yamada S, Tarnawska-Glatt K, Hara-Nishimura I (2020a) Vacuolar processing enzymes in the plant life cycle . New Phytol 226 : 21–31 [ PubMed ] [ Google Scholar ]
- Yamada K, Goto-Yamada S, Nakazaki A, Kunieda T, Kuwata K, Nagano AJ, Nishimura M, Hara-Nishimura I (2020b) Endoplasmic reticulum-derived bodies enable a single-cell chemical defense in Brassicaceae plants . Commun Biol 3 : 21 doi: 10.1038. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yamamoto R, Shinozak K, Masuda Y (1970) Stress-relaxation properties of plant cell walls with special reference to auxin action . Plant Cell Physiol 11 : 947 [ Google Scholar ]
- Yang H, Wang D, Guo L, Ran H, Yvon R, Garman S, Wu HM, Cheung AY (2021) Malectin/Malectin-like domain-containing proteins: a repertoire of cell surface molecules with broad functional potential . Cell Surf 7 : 100056 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites . Plant Cell 17 : 1513–1531 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T , et al. (2018) Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578 : 296–300 [ PubMed ] [ Google Scholar ]
- Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ (2006) Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize . Proc Natl Acad Sci USA 103 : 14664–14671 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yuan M, Shaw PJ, Warn RM, Lloyd CW (1994) Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells . Proc Natl Acad Sci USA 91 : 6050–6053 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Yuan S, Wu Y, Cosgrove DJ (2001) A fungal endoglucanase with plant cell wall extension activity . Plant Physiol 127 : 324–333 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Zhang GF, Staehelin LA (1992) Functional compartmentalization of the Golgi apparatus of plant cells. An immunocytochemical analysis of high pressure frozen and freeze-substituted sycamore maple suspension culture cells . Plant Physiol 99 : 1070–1108 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Zhang L, Huang J, Su S, Wei X, Lin Y, Zhao H, Yu J, Wang J, Hui J, Hao S, et al. (2021a) FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa . Curr Biol 31 : 3004–3016.e4 10.1016/j.cub.2021.04.060 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhang T, Tang H, Vavylonis D, Cosgrove DJ (2019) Disentangling loosening from softening: insights into primary cell wall structure . Plant J 100 : 1101–1117 [ PubMed ] [ Google Scholar ]
- Zhang Y, Yu J, Wang X, Durachko DM, Zhang S, Cosgrove DJ (2021b) Molecular insights into the complex mechanics of plant epidermal cell walls . Science 372 : 706–711 [ PubMed ] [ Google Scholar ]
- Zheng H, Bednarek SY, Sanderfoot AA, Alonso J, Ecker JR, Raikhel NV (2002) NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE . Plant Physiol 129 : 530–539 [ PMC free article ] [ PubMed ] [ Google Scholar ]

Personalize Your Experience
Log in or create an account for a personalized experience based on your selected interests.
Already have an account? Log In
Free standard shipping is valid on orders of $45 or more (after promotions and discounts are applied, regular shipping rates do not qualify as part of the $45 or more) shipped to US addresses only. Not valid on previous purchases or when combined with any other promotional offers.
Register for an enhanced, personalized experience.
Receive free access to exclusive content, a personalized homepage based on your interests, and a weekly newsletter with topics of your choice.
Home / Nutrition & Fitness / 5 common questions about plant-based diets, answered by the experts
5 common questions about plant-based diets, answered by the experts
Please login to bookmark.
Username or Email Address
Remember Me

By now, most people know that prioritizing nutrient-rich fruits, veggies and whole grains is the key to a healthy, well-rounded diet. Meat, on the other hand, tends to be high in saturated fats and can increase your risk of heart attack and stroke , in addition to other health conditions.
For many people, a plant-based diet offers the flexibility needed to maintain healthy, realistic eating over time, while still allowing them to enjoy a range of food groups. However, it can be daunting — and even confusing at times — to make the shift toward plants and away from animal products. This is especially true if you genuinely enjoy the taste of meat or tend to see meat as the “main event” of each meal.
Below, Jason S. Ewoldt, M.S., RDN, LD, a wellness dietitian at Mayo Clinic in Rochester, Minnesota, and part of the Mayo Clinic Healthy Living Program, answers some of the most common questions about plant-based diets, including how to get started.
What is a plant-based diet?
A plant-based diet is a style of eating that prioritizes whole plant foods, such as fruits, vegetables, nuts, beans and legumes, whole grains, and soy products like tofu. Some people eat a plant-based diet and also eat some meat and other animal products, while others don’t.
As a result, “plant based” is often used as an umbrella term and can encompass a range of eating styles and diets, ranging from flexible to more restricted. Vegans and vegetarians, for example, are considered plant-based eaters. They do not eat meat and may avoid other animal products, as well, or in some cases may not consume any animal products. However, it’s perfectly possible to eat meat, dairy and other animal products mindfully and still maintain a plant-based diet. The Mediterranean diet, for example, includes fish and poultry and is often considered one of the healthiest eating styles.
“There are many reasons people opt to go mainly plant based whether they be ethical, religious, health related, personal preference, environmental or otherwise,” says Ewoldt. “Most everyone can benefit from eating more whole-plant foods, regardless of if they include animal products or not.”
What are the pros and cons of a plant-based diet?
Plant-based eating can support your health in a number of ways, says Ewoldt. “Research shows that people on a plant-based diet tend to have the best health outcomes long term with lower rates of heart disease, almost all types of cancer, type 2 diabetes and Alzheimer’s disease, among other chronic diseases.”
Research also indicates that a plant-based diet can reduce erectile dysfunction and vision problems associated with aging, as well as support breast, vaginal and kidney health . Finally, consuming less meat can reduce the risk of stroke, obesity, high blood pressure and high cholesterol.
However, there is a learning curve when it comes to a plant-based diet. “The challenge is that modern food environments are often more conducive to quick, convenient, highly palatable and processed foods rather than whole-plant foods,” says Ewoldt. In other words, not all plant-based foods are made equal. After all, potato chips are technically plant based, but that doesn’t mean eating chips with every meal is a healthy or sustainable option. Often, the switch to a plant-based diet means dedicating more time to planning and cooking your meals and avoiding highly processed foods, regardless of whether or not they are vegan or vegetarian.
Additionally, Ewoldt says it’s important to keep an eye on certain nutrients like protein, vitamin B-12, calcium, iron and vitamin D and ensure that you are getting enough of these nutrients through foods or supplements.
Is it possible to build and maintain muscle by eating a plant-based diet?
Yes, it is possible to both build and maintain muscle on all styles of plant-based eating, including diets that exclude meat.
For adults, the recommended daily intake of protein is around 50 grams a day. With foods like tofu, beans and lentils, Ewoldt says it’s possible to reach the necessary protein intake.
Plants also can offer complete proteins, a kind of protein that contains all nine essential amino acids the human body needs. “To name a few, quinoa, tofu, edamame, Ezekiel bread, hemp seeds and chia seeds are all complete proteins,” says Ewoldt. “Otherwise, you can include noncomplete protein plant foods with other complementary foods to ensure you are getting all of the essential amino acids. Foods like rice and beans, peanut butter on whole-wheat bread, and pita and hummus all form complete proteins.”
Is too much soy bad for hormones?
People are often concerned that soy can increase estrogen levels and the risk of breast cancer — however this isn’t true.
“This misconception happens because soy contains fairly high levels of isoflavones, a type of plant estrogen,” says Ewoldt. “These isoflavones function similarly to human estrogen, however — and here is the important part — with much weaker effects.”
Studies show that soy products, such as tofu and edamame, do not contain enough estrogen to increase the risk of breast cancer. In fact, soy consumption after a breast cancer diagnosis can actually improve cancer outcomes.
How do I get started on a plant-based diet?
The most effective way to start eating a plant-based diet is to lead with simplicity and consistency. Don’t try to overhaul your entire diet overnight.
“If the goal is to eat more plants, a plan needs to be part of the equation,” says Ewoldt. To start, think about your eating habits as a whole, consider areas where you’d like to improve, and identify opportunities to incorporate more nutrient-rich plant foods and recipes .
“Simple ways to start making changes include adding a serving of fruit or vegetables — or both — at every meal, eating fruit as a snack, and including beans in dishes like salads and soups.” In short, most people do best when they focus on what satisfying and tasty foods they can add, rather than what they need to restrict or “forbid.”
It’s also important to remember that food isn’t just about nutrients — it’s also a way people honor their heritage, participate in their culture or religion, connect with family, and make memories. “Making it a point to include plants at each meal can be a realistic approach to eating healthy while still respecting traditions, culture and family,” says Ewoldt. “And keep in mind the goal with healthy eating is not perfection but consistency. There will be times that meals might not be as nutritious as we would like and that’s OK. It’s what we do consistently over time that increases or decreases health, not what we do on occasion.”

Relevant reading
Cook Smart, Eat Well
Healthy, flavorful meals are well within reach for anyone. This book is full of recipes that are easy to put together quickly with basic cooking techniques. Delicious kid friendly recipes, healthy desserts, snacks and side dishes. Being successful in the kitchen doesn’t mean you need to be a gourmet chef. Learn standard food preparation methods and cooking techniques for preparing a wide variety of tasty and healthy meals.

Discover more Nutrition & Fitness content from articles, podcasts, to videos.
You May Also Enjoy

by Jennifer A. Welper, Wellness Executive Chef

by Donald D. Hensrud, M.D., M.S.

by Donald D. Hensrud, M.D., M.S., Jennifer A. Welper, Wellness Executive Chef

Privacy Policy
We've made some updates to our Privacy Policy. Please take a moment to review.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Opportunities and Challenges in Advancing Plant Research with Single-cell Omics
Affiliation.
- 1 Peking University Institute of Advanced Agricultural Sciences, Shandong Laboratory of Advanced Agricultural Sciences in Weifang, Weifang 261325, China.
- PMID: 38996445
- DOI: 10.1093/gpbjnl/qzae026
Plants possess diverse cell types and intricate regulatory mechanisms to adapt to the ever-changing environment of nature. Various strategies have been employed to study cell types and their developmental progressions, including single-cell sequencing methods which provide high-dimensional catalogs to address biological concerns. In recent years, single-cell sequencing technologies in transcriptomics, epigenomics, proteomics, metabolomics, and spatial transcriptomics have been increasingly used in plant science to reveal intricate biological relationships at the single-cell level. However, the application of single-cell technologies to plants is more limited due to the challenges posed by cell structure. This review outlines the advancements in single-cell omics technologies, their implications in plant systems, future research applications, and the challenges of single-cell omics in plant systems.
Keywords: Multi-omics; Single-cell epigenomics; Single-cell proteomics; Single-cell transcriptomics; Spatial transcriptomics.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
PubMed Disclaimer
Similar articles
- Advances and applications of single-cell omics technologies in plant research. Mo Y, Jiao Y. Mo Y, et al. Plant J. 2022 Jun;110(6):1551-1563. doi: 10.1111/tpj.15772. Epub 2022 Apr 28. Plant J. 2022. PMID: 35426954 Review.
- Defining Cell Identity with Single-Cell Omics. Mincarelli L, Lister A, Lipscombe J, Macaulay IC. Mincarelli L, et al. Proteomics. 2018 Sep;18(18):e1700312. doi: 10.1002/pmic.201700312. Epub 2018 May 28. Proteomics. 2018. PMID: 29644800 Free PMC article. Review.
- Single-Cell Omics Analyses Enabled by Microchip Technologies. Deng Y, Finck A, Fan R. Deng Y, et al. Annu Rev Biomed Eng. 2019 Jun 4;21:365-393. doi: 10.1146/annurev-bioeng-060418-052538. Epub 2019 Mar 18. Annu Rev Biomed Eng. 2019. PMID: 30883211 Review.
- Single-cell omics in ageing: a young and growing field. He X, Memczak S, Qu J, Belmonte JCI, Liu GH. He X, et al. Nat Metab. 2020 Apr;2(4):293-302. doi: 10.1038/s42255-020-0196-7. Epub 2020 Apr 20. Nat Metab. 2020. PMID: 32694606 Review.
- Advanced Omics Techniques for Understanding Cochlear Genome, Epigenome, and Transcriptome in Health and Disease. Tisi A, Palaniappan S, Maccarrone M. Tisi A, et al. Biomolecules. 2023 Oct 17;13(10):1534. doi: 10.3390/biom13101534. Biomolecules. 2023. PMID: 37892216 Free PMC article. Review.
Publication types
- Search in MeSH
Related information
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
Which is a new interdisciplinary subject meant to store and organize biological data? A. Biochemistry B. Biotechnology C. Bioengineering D. Bioinformatics
Bioinformatics is correct answer.
Explanation:
Yea, any questions?
ask me then.
Related Questions
When this cell divides to form two cells, which diagram shows the most likely result?
I am 90% sure it is a
A microscope has a low-power magnification of 200x and a high-power magnification of 1600x. If the low-power field diameter is 1.2 mm, calculate the high power field diameter in micrometers
The diameter of the high power field in micrometers is 9600 micrometer (9.6 mm).
[tex]\frac{200}{1600} =\frac{1,2}{x}[/tex] = 9,6 mm
Compound microscope lenses are called objective lens and ocular lens . The objective lens is a lens that is installed very close to the object being observed. The ocular lens is very close to the observer's eye .
With this information, we can conclude that the high power field in micrometers is 9600 micrometer (9.6 mm).
Learn more about microscope in brainly.com/question/18661784
what is your favorite spongebob character? trying to get rid of points.
Which part of the scientific method is best illustrated by the biologist wondering why there were so few frogs in the second year compared to the first? O forming a hypothesis O making an observation and asking a question O collecting data O setting up an experiment
I think it is making an observation and asking a question
Scientific methods are the procedural steps that involve experimentation to reach conclusion. The method best represents the observations and asking of questions . Thus, option B is correct.
The scientific method has been defined as the multistep process that involves various method procedures, including the questions, research topic issues, hypothesis , experimental and research tests, analysis of data, and drawing of conclusions .
The scientific method primarily starts with a question about the subject matter that needs to be solved for which the research is performed and a hypothesis is given. After a hypothesis, is established experiments are conducted so as to make observations.
The observations are then analyzed to draw conclusions to present the findings. This interpretation gives the solution to the presented problems. The biologist was asking questions when he observed and compared the population of the frogs in the first and second years.
Therefore, in option B. the biologist asked questions based on the observations .
Learn more about the scientific method, here:
https://brainly.com/question/16172865
what happens when iodine is added to soaked maize grain overnight. observation and interference
Have students mix water and cornstarch in a cup and then add iodine solution to it. The solution will turn a dark purple. This will show that iodine, when in the presence of starch, turns purple, the basis of the other two experiments.
Enzymes Which of the following can be cofactors?
Cofactors can either be ions, such as zinc and iron ions, or organic molecules, such as vitamins or vitamin-derived molecules. Many of these cofactors will attach near the substrate binding site to facilitate the binding of the substrate to the enzyme.
Cells are limited in size because Select one: O a. they divide before they have a chance to grow O b. the cell membrane cannot supply enough materials for a cell that is too large O c. O d. they are always in groups they do not have enough vacuoles to store materials
because cell are made up of many other tissue and they do not have enough vacuoles to store materials
What does a food web show? A. how much energy gets transferred between trophic levels B. the complex interactions and feeding relationships between organisms within an ecosystem C. the cycle of organic materials in an ecosystem PLEASE HELP
The answer is B because A is an energy pyramid, and it cannot be C because a food web is not a cycle as it doesn't restart.
what effect does the increased temperature have on the ecology and distribution of plant species
Higher temperature affects plants’ growth especially in temperate crops like wheat. It reduces the growth of shoots and roots in plants. This, in turn, results in a substantial reduction in the overall height of the plant. High air temperature can lead to dehydration and scorched leaves .
In general, the increase of temperature would cause a damage in ecology of a certain area as most plants would not be able to adapt to this change. This, in turn, will affect the distribution of such plant species as they would die off unless placed in an environment with more favorable conditions .
The impact of increased temperature on plant ecology and distribution can lead to shifts in species ranges, altered species interactions, changes in ecosystem functions, and challenges in adapting to new climatic conditions.
Some of the key impacts are as follows:
Thus, increased temperature's effects on plant ecology and distribution can modify the ranges of various species, alter how they interact with one another, alter how ecosystems work, and make it more difficult for them to adapt to changing climatic conditions.
To know more about Ecology , click here:
https://brainly.com/question/13046612
¿What is Bacteria? ..
bacteria are mostly free-living organisms often consisting of one biological cell. This means they are unicellular organisms, unlike humans, who are composed of millions of cells. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, and the deep biosphere of Earth's crust.
Hope this helps!
[tex]\sf{\qquad\qquad\huge\underline{{\sf Answer}}}[/tex]
¿ What is Bacteria?
They are the primitive beings that inhabit the planet earth, prokaryotic microorganisms , not visible to the nak - ed eye.
They lack organelles and a nuclear membrane, their genetic material is scattered in the cytoplasm.
There are 3 types of Bacteria which are :-
I hope I've helped : )
what role we can play to manage solid waste
To manage solid waste, the steps we can take are :
Don’t throw chocolate, biscuit, ice cream, and other fast-food material wrappers on roads or in open spaces. Keep them in the pocket and put them in a specific bucket later on.
As alternatives to plastic bags, we can use cloth bags made of household material. i.e old strains, curtain material, bed sheets, etc.
Avoid the Avoid use of tissue paper instead of using your napkin.
Social Awareness programs can be organized programs communication ,schools and colleges.
To encourage the citizens to participate in waste management.
Avoid the use of tissue paper; instead, use your napkin.
Social Awareness programs can be organized in communities, schools, and colleges.
Please mark it as brainliest and I hope it helps:)
I'm not good at this.Can someone solve this?
A person who has type AB blood and is heterozygous for Rh factor has children with a person who is heterozygous for type A blood and is negative for Rh factor. List all possible gametes these two individuals could produce and list the genotype and phenotype ratios of the F1 generation.
Blood type is determined by the alleles coding for the antigen present on cell membranes.
The person with type AB blood has a heterozygou s Rh factor.
The person with heterozygous type A blood is negative for Rh factor
The genotype ratio is 1:1:1:1:1:1:1:1
The phenotype ratio is 2: 2:1:1:1:1.
Learn more about blood types here:
https://brainly.com/question/21209477
Whats the correct answer answer asap for brainlist
How do results prove the fish are genetically modified? If both parents are fluorescent, can they produce a regular-colored offspring? What conclusions came from this experiment?
The results show that fish are GMO when they fluoresce at night, GFP fish produce colored offspring when germinal cells are modified and conclusions indicate that transgenesis is viable.
The Green fluorescent protein (GFP) is a protein used in molecular biology to produce genetically modified organisms ( GMOs ).
The GFP gene can produce stable fluorescent offspring if the transgenesis technique modifed the germinal line.
In conclusion, we can conclude that fish were genetically modified because they express the GFP protein and produce fluorescent offspring only when the germinal line was modified.
Learn more about the GFP protein here:
https://brainly.com/question/15236303
Answer asap whats the correct answer for brainlist
Answer: The fertile soil, it limited the number of Japanese who could earn a living from farming
What would happen if all the plants on earth died?
Answer:Without plants there would no longer be oxygen for animals to use to respire therefore animals will die.
And herbivores animals will no longer be able to feed on plants and they will also die.
Which type of cells have cell membrane
Answer: animal cells
plant cells have a wall
What part of the body uses active transport to take up nutrients after concentration has reached equilibrium
The villi of the small intestine use active transport to take up nutrients after concentration has reached equilibrium.
Active transport may be defined as the process that occurs against the concentration gradient and is mediated by carrier proteins. Metabolic energy is used to move ions or molecules against a concentration gradient.
During the process of digestion, the villi in the small intestine enthrall the soluble nutrients gradually. Over time, the concentration of nutrients in the villi acquires an equilibrium with the concentration in the gut. Until here, the nutrient uptake is carried by the process of passive diffusion .
But after attaining the equilibrium , the nutrient uptake is carried by the process of active transport .
The complete question is as follows:
What part of the body uses active transport to take up nutrients after concentration has reached equilibrium?
Therefore, the correct option for this question is C , i.e. small intestine .
To learn more about Active transport , refer to the link:
https://brainly.com/question/18434867
Using the following diagram, which image shows the absorption of the highest energy photon?
When electrons absorb energy, they move to higher orbital levels. The highest energy levels are the furthest from the nucleus. More energy is needed to occupy higher energy levels . Therefore, the electron which must have absorbed the most energy is in Diagram D. In this diagram, the electron absorbs enough energy to move it from n = 1 to n = 4.
When electrons absorb energy, they move to higher energy levels. In the given question, option D shows maximum energy absorption as electrons move from n = 1 to n = 4 . Thus, the correct option is D .
Electron transition is the transition of an electron from one energy level to another within an atom or artificial atom. In this, the electron "jumps" from one energy level to another, typically in a few nanoseconds or less. This transition is favored by the difference in energy in different energy levels.
When an electron absorbs energy , they move to higher energy orbital levels. The highest energy levels are the ones farthest from the nucleus and the lowest energy levels are the ones that are nearest to the nucleus . As more energy is required to stay at higher energy levels. Therefore, the electron must absorb the most energy.
In this diagram, option D denotes the highest energy photon as the electron absorbs enough energy to move from n = 1 to n = 4 .
Therefore, the correct option is D .
Learn more about photons here:
https://brainly.com/question/20912241
Consider this animal cell. the organelles in an animal cell are labeled. part e represents small dots on the nucleolus. what is the function of the small, dark organelles labeled e?
Part e represents small dots on the nucleolus. The function of the small, dark organelles labeled e is that they make proteins for the cell. hence the small dots represent Ribosomes .
learn more about ribosomes here: https://brainly.com/question/8773679
Fish populations and other aquatic resources are likely to be affected by changes in what aquatic factors? a. water temperatures b. disease c. water quality d. all of the above please select the best answer from the choices provided a b c d
Fish populations and other aquatic resources are likely to be affected by changes in all of the above i.e., water temperatures, disease and water quality.
a) Water temperatures: Warm water released from industries into the rivers has lesser BOD (biological oxygen demand) than cool water. The marine species which are unable to migrate will die due to the unavailability of oxygen while frequent algal blooms will be present due to no availability of oxygen.
b) Diseases : Increase in toxins and chemicals will increase in the river as a result of the industrial dump. This will cause industrial animals to acquire toxicants in their bodies and will cause several diseases , wounds, and eventually the death of the organism.
c) Water quality: due to warm and toxic water many marine animals will migrate to places with cooler water. there will be increased algal blooms. This will affect the whole food web and hence there will be a loss of biodiversity.
hence all of the above factors will affect Fish populations and other aquatic resources.
learn more about Fish populations here: https://brainly.com/question/1235358
¿What are invertebrates?
¿What are invertebrates?
Invertebrates are unicellular or multicellular organisms that present a great variety of habits and morpho-functional patterns.
Which are classified into : -
Anthropods , molluscs, worms, echinoderms, coelenterates , and porifera .
What is the difference between a Triglyceride and a Phospholipid? A. Triglycerides have 2 fatty acid chains, Phospholipids have 3. B. Triglycerides have a phosphate group, Phospholipids do not. C. There is no difference, they are the same thing. D. Phospholipids have a phosphate group, Triglycerides do not.
A. Triglycerides have 2 fatty acid chains, Phospholipids have 3.
The main difference between the structures of phospholipid and triglycerides is that a phospholipid molecule is made up of a glycerol attached to two fatty acids and a phosphate group while on the other hand, triglyceride is composed of three fatty acid groups.
So we can conclude that option A is the correct answer.
Learn more bout triglycerides here: https://brainly.com/question/5028914
The main difference between a Triglyceride and a Phospholipid is that Phospholipids have a phosphate group, while Triglycerides do not (Option D).
A Phospholipid is a lipidic molecule that contains a phosphate group, which represents the main component of the cell membrane.
Conversely, Triglycerides are lipids composed of three fatty acid chains linked to a glycerol backbone.
In conclusion, Phospholipids have a phosphate group, while Triglycerides do not (Option D).
Learn more about triglycerides here:
https://brainly.com/question/5028914
What are the three measures of life’s variations?
What are three measures of life variations?
Species diversity, genetic diversity, ecosystem diversity
Biodiversity
The amount of biological or living diversity per unit area. It includes the concepts of species diversity, habitat diversity and genetic diversity.
I hope this helps and have a great day!
What five intermolecular forces of attraction are important for tertiary structure?
for protein the force of attractions for tertiary structure are
(i) ionic bonds
(ii) hydrogen bonds
(iii) van der Waals forces and electrostatic bonds
(iv) disulphide linkages
(v) Hydrophobic Interactions.
Why do organisms have varying traits, like different colored eggs? What might drive change in egg color in natural population? Is this experiment an example of natural or artificial selection and why?
the organisms have varying traits, like different colored eggs because of the natural selection .
Natural selection is one of the main mechanisms of evolution . In a very simple way, we can say that natural selection is a process in which the fittest organisms are selected, survive in the environment , reproduce and pass their characteristics on to their descendants .
the color change of an egg occurs by natural selection and also by genetic variability . since the environment will select the best ones to survive and mutations will occur. Natural selection will forcefully change the environment causing only a few species to survive .
See more about natural selection at brainly.com/question/9830102
What if the surface of a bacteria cell was not very antigenic? What would this mean for the host
If the surface of a bacteria cell was not very antigenic then it can enter host cells and eventually cause harm.
An antigen is any molecular structure present in a pathogenic organism (e.g., bacteria) that can be detected by the immune system.
Antigens usually bind to antibodies that are able to bind specific antigenic features and thus trigger a concomitant immune response.
In conclusion, if a bacteria cell was not very antigenic then it can cause harm.
Learn more about antigens here:
https://brainly.com/question/7597406
Question 13 of 20: Select the best answer for the question. 13. Which of the following is formed when a group of tissues works together to perform a common function? A. Organ system B. Organ C. Body D. Brain Mark for review (Will be highlighted on the review page) << Previous Question 9,269 JUL 18
the following is formed when a group of tissues works together to perform a common function is called an Organ.
The word organ derives from the Latin term organum (instrument, tool). These structures are objects of physiology , the area of biology that studies how organisms, systems, organs, cells and molecules perform chemical and physical functions in the body.
In this case, is correct to say that an organ is formed when a group of tissues works together to perform a common function.
See more about organ at brainly.com/question/12825206
In which organizational level of life does natural selection occur? A. Tissues B. Ecosystems C. Populations D. Organisms
It happens in populations

IMAGES
VIDEO
COMMENTS
Regeneration of a whole plant from a single leaf mesophyll protoplast. A, A protoplast isolated from a leaf mesophyll cell. B, Protoplast cells that have undergone several rounds of cell division. C, Shoot meristem formation from protoplast-derived callus. D, A regenerated plant. Arrowheads in (A) mark a chloroplast.
Expert commentary on major open questions in photosynthesis research. Nancy A Eckardt, Yagut Allahverdiyeva, Clarisa E Alvarez, Claudia Büchel, Adrien Burlacot, Tanai Cardona, Emma Chaloner, Benjamin D Engel, Arthur R Grossman, Dvir Harris, Nicolas Herrmann, Michael Hodges, Jan Kern, Tom Dongmin Kim, Veronica G Maurino, Conrad W Mullineaux, Henna Mustila, Lauri Nikkanen, Gabriela Schlau-Cohen ...
Affiliations 1 Weill Institute for Cell and Molecular Biology and School of Integrative Plant Science, Section of Plant Biology, Cornell University, Ithaca, New York 14853, USA.; 2 Department of Botany and Center for Quantitative Cell Imaging, University of Wisconsin-Madison, Wisconsin 53706, USA.; 3 Department of Biology and Center for Engineering Mechanobiology, Washington University in St ...
Science happens in the interface between known and unknown. In the review article Fifteen compelling open questions in plant cell biology, published in The Plant Cell Focus Issue on Plant Cell Biology, we focus on the unknown, the mysteries waiting to be solved. We asked fifteen experts to write a short description of the biggest open question ...
Another important unanswered question concerns the function of the diverse hydroxyproline rich glycoprotein (HRGP) polymers. The recent findings that most RG-I in the wall appears to be covalently attached to AGPs ( Tan et al., 2023) and that Leucine Rich Repeat eXtensin-Rapid ALkalinization Factor (LRX-RALF) complexes structure the cell wall ...
In this Focus Issue, we go back to the "roots" of the journal, that gave it its name, and turn our attention to the many fascinating facets of plant cell biology. The 11 review papers and 11 original research articles in this issue exemplify the breadth of the field and highlight both established concepts and recent achievements.
Abstract. As scientists, we are at least as excited about the open questions-the things we do not know-as the discoveries. Here, we asked 15 experts to describe the most compelling open questions in plant cell biology. These are their questions: How are organelle identity, domains, and boundaries maintained under the continuous flux of vesicle ...
The Plant Cell, Volume 35, Issue 1, January 2023, Pages ... We present unresolved questions in plant abiotic stress biology as posed by 15 research groups with expertise spanning eco-physiology to cell and molecular biology. ... An important question for research is whether these trade-offs, or other ecologically important tradeoffs, can be ...
As scientists, we are at least as excited about the open questions—the things we do not know—as the discoveries. Here, we asked 15 experts to describe the most compelling open questions in ...
A network of demethylesterified homogalacturonan filaments formed by a pectin-binding LRX8-RALF4 complex was observed in pollen tube cell walls ( Moussu et al., 2023, Moussu and Ingram, 2023 ). This complex exerts a condensing effect, thus patterning the cell wall polymers into a reticulated network critical for cell wall integrity and ...
For the purposes of this paper, the panel decided to categorize the questions into five broad areas that reflect the breadth and depth of plant research discussed during the 2-d workshop: Society, Environment and adaptation, Species interactions, Understanding and utilizing plant cells, and Diversity.
R35 GM136637/GM/NIGMS NIH HHS/United States. In this glossary of plant cell structures, we asked experts to summarize a present-day view of plant organelles and structures, including a discussion of outstanding questions. In the following short reviews, the authors discuss the complexities of the plant cell endomembrane system, exciting connec ….
Plant cell biology is the study of all aspects of plant cells. It is particularly concerned with structure, growth, division, signalling, differentiation and death of plant cells. Proteolysis in ...
Question. 3 answers. Nov 19, 2017. if there are many copies of gene on DNA of plant cell as a result of evolution of a specific gene .and as a result the expression of that gene increase and ...
Introduction. In 2011, we published 100 important questions for plant science research pertaining to (A) society, (B) environment and adaptation, (C) species interactions, (D) understanding and utilising plant cells, and (E) diversity (Grierson et al., 2011).The original publication became one of the most read articles in New Phytologist, demonstrating wide interest in identifying priorities ...
An intriguing and still largely unexplored question in plant cell wall research is how composition and structure change with developmental stage progression. Changes occurring on the time scale of minutes to days have recently been investigated due to advances in live-imaging technologies, such as microfluidics, and non-invasive techniques ...
Plant cell wall researchers were asked their view on what the major unanswered questions are in their field. This article summarises the feedback that was received from them in five questions. ... Top five unanswered questions in plant cell surface research Cell Surf. 2024 Feb 13:11:100121. doi: 10.1016/j.tcsw.2024.100121. eCollection 2024 Jun ...
Aug 15, 2023. Answer. Plants have the greatest net production efficiencies, which range from 30-85%. The reason that some organisms have such low net production efficiencies is that they are ...
We sought questions about plants, plant science and plant research from diverse sources, ranging from the botanically curious nonexpert to those with years of plant-focussed research expertise. Our panel of experts was chosen to reflect the diverse interests of plant research around the world, with 10 each from both the Global North and South.
Explore a collection of the most read and most cited articles making an impact in The Plant Cell published within the past two years. This collec ... High-Impact Research from The Plant Cell. ... We present unresolved questions in plant abiotic stress biology as posed by 15 research groups with expertise spanning eco-physiology to cell and ...
abstract = "Plant cell wall researchers were asked their view on what the major unanswered questions are in their field. This article summarises the feedback that was received from them in five questions.
Interactions between plants and their microbiome are bidirectional. With a belowground bias, we first focus on the processes leading to root and rhizosphere microbiome assembly, inspecting first the plant component ( in Figure 1) of the 'plant-to-microbiome' direction.In this component, the key research question is 'How do plants impact their microbiome?'.
The field of plant cell biology has a rich history of discovery, going back to Robert Hooke's discovery of cells themselves. The development of microscopes and preparation techniques has allowed for the visualization of subcellular structures, and the use of protein biochemistry, genetics, and molecular biology has enabled the identification of proteins and mechanisms that regulate key ...
Open questions in plant cell wall synthesis. March 2023. Journal of Experimental Botany 74 (12) DOI: 10.1093/jxb/erad110. Authors: Heather E McFarlane. To read the full-text of this research, you ...
By now, most people know that prioritizing nutrient-rich fruits, veggies and whole grains is the key to a healthy, well-rounded diet. Meat, on the other hand, tends to be high in saturated fats and can increase your risk of heart attack and stroke, in addition to other health conditions.. For many people, a plant-based diet offers the flexibility needed to maintain healthy, realistic eating ...
However, the application of single-cell technologies to plants is more limited due to the challenges posed by cell structure. This review outlines the advancements in single-cell omics technologies, their implications in plant systems, future research applications, and the challenges of single-cell omics in plant systems.
The main difference between a Triglyceride and a Phospholipid is that Phospholipids have a phosphate group, while Triglycerides do not (Option D).. What is a Phospholipid? A Phospholipid is a lipidic molecule that contains a phosphate group, which represents the main component of the cell membrane.. Conversely, Triglycerides are lipids composed of three fatty acid chains linked to a glycerol ...